Normalization of clonal diversity in gene therapy studies using shape constrained splines
- PMID: 35264585
- PMCID: PMC8907296
- DOI: 10.1038/s41598-022-05837-0
Normalization of clonal diversity in gene therapy studies using shape constrained splines
Abstract
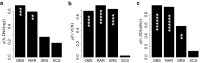
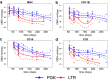
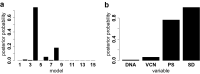
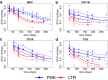
Viral vectors are used to insert genetic material into semirandom genomic positions of hematopoietic stem cells which, after reinfusion into patients, regenerate the entire hematopoietic system. Hematopoietic cells originating from genetically modified stem cells will harbor insertions in specific genomic positions called integration sites, which represent unique genetic marks of clonal identity. Therefore, the analysis of vector integration sites present in the genomic DNA of circulating cells allows to determine the number of clones in the blood ecosystem. Shannon diversity index is adopted to evaluate the heterogeneity of the transduced population of gene corrected cells. However, this measure can be affected by several technical variables such as the DNA amount used and the sequencing depth of the library analyzed and therefore the comparison across samples may be affected by these confounding factors. We developed an advanced spline-regression approach that leverages on confounding effects to provide a normalized entropy index. Our proposed method was first validated and compared with two state of the art approaches in a specifically designed in vitro assay. Subsequently our approach allowed to observe the expected impact of vector genotoxicity on entropy level decay in an in vivo model of hematopoietic stem cell gene therapy based on tumor prone mice.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

