A non-canonical tricarboxylic acid cycle underlies cellular identity
- PMID: 35264789
- PMCID: PMC8934290
- DOI: 10.1038/s41586-022-04475-w
A non-canonical tricarboxylic acid cycle underlies cellular identity
Abstract
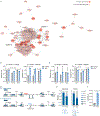
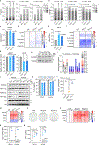
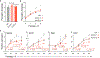
The tricarboxylic acid (TCA) cycle is a central hub of cellular metabolism, oxidizing nutrients to generate reducing equivalents for energy production and critical metabolites for biosynthetic reactions. Despite the importance of the products of the TCA cycle for cell viability and proliferation, mammalian cells display diversity in TCA-cycle activity1,2. How this diversity is achieved, and whether it is critical for establishing cell fate, remains poorly understood. Here we identify a non-canonical TCA cycle that is required for changes in cell state. Genetic co-essentiality mapping revealed a cluster of genes that is sufficient to compose a biochemical alternative to the canonical TCA cycle, wherein mitochondrially derived citrate exported to the cytoplasm is metabolized by ATP citrate lyase, ultimately regenerating mitochondrial oxaloacetate to complete this non-canonical TCA cycle. Manipulating the expression of ATP citrate lyase or the canonical TCA-cycle enzyme aconitase 2 in mouse myoblasts and embryonic stem cells revealed that changes in the configuration of the TCA cycle accompany cell fate transitions. During exit from pluripotency, embryonic stem cells switch from canonical to non-canonical TCA-cycle metabolism. Accordingly, blocking the non-canonical TCA cycle prevents cells from exiting pluripotency. These results establish a context-dependent alternative to the traditional TCA cycle and reveal that appropriate TCA-cycle engagement is required for changes in cell state.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures

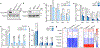

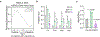


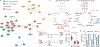



Comment in
-
Alternative cycle for citrate.Nat Rev Mol Cell Biol. 2022 May;23(5):305. doi: 10.1038/s41580-022-00475-w. Nat Rev Mol Cell Biol. 2022. PMID: 35296797 No abstract available.
-
Krebs and an alternative TCA cycle!Cell Res. 2022 Jun;32(6):509-510. doi: 10.1038/s41422-022-00664-4. Cell Res. 2022. PMID: 35459937 Free PMC article. No abstract available.
-
Krebs takes a turn at cell differentiation.Cell Metab. 2022 May 3;34(5):658-660. doi: 10.1016/j.cmet.2022.04.005. Cell Metab. 2022. PMID: 35508108
-
Light shed on a non-canonical TCA cycle: cell state regulation beyond mitochondrial energy production.Signal Transduct Target Ther. 2022 Jun 28;7(1):201. doi: 10.1038/s41392-022-01060-5. Signal Transduct Target Ther. 2022. PMID: 35764605 Free PMC article. No abstract available.
-
Identification of metabolic networks by genetic co-essentiality analysis.Nat Rev Mol Cell Biol. 2023 Jun;24(6):378. doi: 10.1038/s41580-023-00608-9. Nat Rev Mol Cell Biol. 2023. PMID: 37101011 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

