PlasticDB: a database of microorganisms and proteins linked to plastic biodegradation
- PMID: 35266524
- PMCID: PMC9216477
- DOI: 10.1093/database/baac008
PlasticDB: a database of microorganisms and proteins linked to plastic biodegradation
Abstract
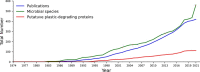
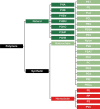
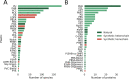
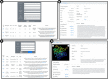
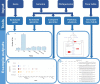
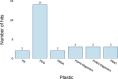
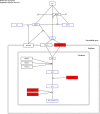
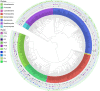
The number of publications reporting putative plastic-degrading microbes and proteins is continuously increasing, necessitating the compilation of these data and the development of tools to facilitate their analysis. We developed the PlasticDB web application to address this need, which comprises a database of microorganisms and proteins reported to biodegrade plastics. Associated metadata, such as the techniques utilized to assess biodegradation, the environmental source of microbial isolate and presumed thermophilic traits are also reported. Proteins in the database are categorized according to the plastic type they are reported to degrade. Each protein structure has been predicted in silico and can be visualized or downloaded for further investigation. In addition to standard database functionalities, such as searching, filtering and retrieving database records, we implemented several analytical tools that accept inputs, including gene, genome, metagenome, transcriptomes, metatranscriptomes and taxa table data. Users can now analyze their datasets for the presence of putative plastic-degrading species and potential plastic-degrading proteins and pathways from those species. Database URL:http://plasticdb.org.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
-
- Siddiqui J. and Pandey G. (2013) A review of plastic waste management strategies. Int. Res. J. Environ. Sci., 2, 84–88.
-
- Lebreton L. and Andrady A. (2019) Future scenarios of global plastic waste generation and disposal. Palgrave Commun., 5, 1–11.
-
- Zantis L.J., Carroll E.L., Nelms S.E. et al. (2021) Marine mammals and microplastics: a systematic review and call for standardisation. Environ. Pollut., 269, 116142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

