Acids produced by lactobacilli inhibit the growth of commensal Lachnospiraceae and S24-7 bacteria
- PMID: 35266847
- PMCID: PMC8920129
- DOI: 10.1080/19490976.2022.2046452
Acids produced by lactobacilli inhibit the growth of commensal Lachnospiraceae and S24-7 bacteria
Abstract
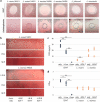
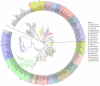
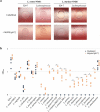
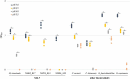
The Lactobacillaceae are an intensively studied family of bacteria widely used in fermented food and probiotics, and many are native to the gut and vaginal microbiota of humans and other animals. Various studies have shown that specific Lactobacillaceae species produce metabolites that can inhibit the colonization of fungal and bacterial pathogens, but less is known about how Lactobacillaceae affect individual bacterial species in the endogenous animal microbiota. Here, we show that numerous Lactobacillaceae species inhibit the growth of the Lachnospiraceae family and the S24-7 group, two dominant clades of bacteria within the gut. We demonstrate that inhibitory activity is a property common to homofermentative Lactobacillaceae species, but not to species that use heterofermentative metabolism. We observe that homofermentative Lactobacillaceae species robustly acidify their environment, and that acidification alone is sufficient to inhibit growth of Lachnospiraceae and S24-7 growth, but not related species from the Clostridiales or Bacteroidales orders. This study represents one of the first in-depth explorations of the dynamic between Lactobacillaceae species and commensal intestinal bacteria, and contributes valuable insight toward deconvoluting their interactions within the gut microbial ecosystem.
Keywords: Bacteroidales; Clostridiales; Lachnospiraceae; Muribaculaceae; Probiotics; S24-7; acid stress; gut; lactic acid bacteria; lactobacilli; microbiota.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


References
-
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document: the International scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–514. doi:10.1038/nrgastro.2014.66. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
