Hyperinflammatory Syndrome, Natural Killer Cell Function, and Genetic Polymorphisms in the Pathogenesis of Severe Dengue
- PMID: 35267010
- PMCID: PMC9574659
- DOI: 10.1093/infdis/jiac093
Hyperinflammatory Syndrome, Natural Killer Cell Function, and Genetic Polymorphisms in the Pathogenesis of Severe Dengue
Abstract
Background: Severe dengue, characterized by shock and organ dysfunction, is driven by an excessive host immune response. We investigated the role of hyperinflammation in dengue pathogenesis.
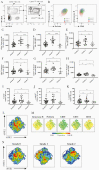
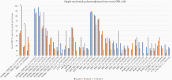
Methods: Patients recruited into an observational study were divided into 3 plasma leak severity grades. Hyperinflammatory biomarkers were measured at 4 time points. Frequencies, activation, and cytotoxic potential of natural killer (NK) cells were analyzed by flow cytometry. RNA was extracted from sorted CD56+ NK cells and libraries were prepared using SMART-Seq and sequenced using HiSeq3000 (Illumina).
Results: Sixty-nine patients were included (grade 0, 42 patients; grade 1, 19 patients; grade 2, 8 patients). Patients with grade 2 leakage had higher biomarkers than grade 0, including higher peak ferritin levels (83.3% vs 45.2%) and H-scores (median, 148.5 vs 105.5). NK cells from grade 2 patients exhibited decreased expression of perforin and granzyme B and activation markers. RNA sequencing revealed 3 single-nucleotide polymorphisms in NK cell functional genes associated with more severe leakage-NK cell lectin-like receptor K1 gene (KLRK1) and perforin 1 (PRF1).
Conclusions: Features of hyperinflammation are associated with dengue severity, including higher biomarkers, impaired NK cell function, and polymorphisms in NK cell cytolytic function genes (KLRK1 and PRF1). Trials of immunomodulatory therapy in these patients is now warranted.
Keywords: dengue; hyperinflammation; immunomodulation; macrophage activation syndrome; single-nucleotide polymorphisms.
© The Author(s) 2022. Published by Oxford University Press for the Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. S. Y. reports receiving personal fees as a member of the Roche advisory board on severe dengue and for work on the Janssen Pharmaceuticals advisory board for dengue antiviral development. All others authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures

References
-
- Cattarino L, Rodriguez-Barraquer I, Imai N, Cummings DAT, Ferguson NM.. Mapping global variation in dengue transmission intensity. Sci Transl Med 2020; 12:eaax4144. - PubMed
-
- Yacoub S, Mongkolsapaya J, Screaton G.. Recent advances in understanding dengue. F1000Res 2016; 5:F1000 Faculty Rev-78.
-
- Screaton G, Mongkolsapaya J, Yacoub S, Roberts C.. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol 2015; 15:745–59. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

