Evaluation of the Clinical Utility of Genomic Profiling to Inform Selection of Clinical Trial Therapy in Salivary Gland Cancer
- PMID: 35267442
- PMCID: PMC8909363
- DOI: 10.3390/cancers14051133
Evaluation of the Clinical Utility of Genomic Profiling to Inform Selection of Clinical Trial Therapy in Salivary Gland Cancer
Abstract
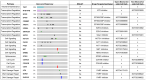
For most patients with salivary gland cancer, there are no effective standard systemic therapies. Although clinical trials of biomarker-led drug therapies have delivered significant recent advances, there remains a need to understand the clinical utility of genomic profiling of cancer as a means to match patients with recurrent or metastatic salivary gland cancer to clinical trial therapies. In total, 209 patients with salivary gland cancers were profiled with 24 gene (n = 209)) and >325 gene (n = 32) DNA-based next-generation sequencing panels. A retrospective systematic evaluation was performed to identify the frequency of available matched drug therapies within clinical trials based on the results. The matches were then stratified based upon the level of evidence supporting the drug−biomarker combination being investigated using the ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT) to determine the strength of the clinical rationale for each gene−drug match identified. DNA-based next generation sequencing (NGS) analysis was successful in 175/209 (84%) patients with salivary gland cancer. Using the 24-gene NGS panel, actionable alterations were identified in 27% (48/175) patients. Alterations were most frequent in salivary duct carcinoma (88%) characterized by TP53 and/or PIK3CA mutations, with matched trials available for 63% (10/16). In ACC, biomarker-matched trials were available for 7% (8/115), and no genomic alterations were found in 96/115 (83%) of ACC patients. TP53 was the most frequently altered gene across all subtypes; however, there were no trials recruiting based on TP53 status. In 32 ACC patients with no genomic alterations using the 24-gene panel, a broader (>325 gene) panel identified alterations in 87% (27/32) of cases with biomarker-matched trials available in 40% (13/32) cases. This study identified that genomic profiling using focused (24-gene) NGS panels has potential utility in matching to trial therapies for most patients with non-ACC salivary gland cancer. For patients with ACC, broader genomic profiling has demonstrated added clinical utility. We describe the application of an approach to classification of levels of evidence which may be helpful to inform the clinician and patient decision making around the selection of clinical trial therapies.
Keywords: adenoid cystic carcinoma; biomarker; clinical trials; molecular profiling; salivary duct carcinoma; salivary gland cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Doebele R.C., Drilon A., Paz-Ares L., Siena S., Shaw A.T., Farago A.F., Blakely C.M., Seto T., Cho B.C., Tosi D., et al. Entrectinib in Patients with Advanced or Metastatic NTRK Fusion-Positive Solid Tumours: Integrated Analysis of Three Phase 1-2 Trials. Lancet Oncol. 2020;21:271–282. doi: 10.1016/S1470-2045(19)30691-6. - DOI - PMC - PubMed
-
- Drilon A., Laetsch T.W., Kummar S., DuBois S.G., Lassen U.N., Demetri G.D., Nathenson M., Doebele R.C., Farago A.F., Pappo A.S., et al. Efficacy of Larotrectinib in TRK Fusion–Positive Cancers in Adults and Children. N. Engl. J. Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

