Role of Neural (N)-Cadherin in Breast Cancer Cell Stemness and Dormancy in the Bone Microenvironment
- PMID: 35267624
- PMCID: PMC8909418
- DOI: 10.3390/cancers14051317
Role of Neural (N)-Cadherin in Breast Cancer Cell Stemness and Dormancy in the Bone Microenvironment
Abstract
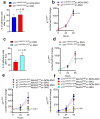
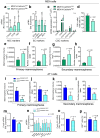
Breast cancer cells that interact with spindle-shaped N-Cadherin+ Osteoblasts (SNOs) are recognised to become dormant through a Notch2-dependent mechanism. We found that Notch2High human BrCa MDA-MB231 (MDA) cells also expressed high level of N-Cadherin. This prompted us to hypothesize that N-Cadherin could have a role in MDA-SNO interaction. Of note, the expression of N-Cadherin in MDA cells reduced tumour incidence and bone osteolysis in BrCa mouse model. Moreover, similarly to Notch2High MDA cells, the N-CadherinHigh MDA cells revealed a high expression of the canonical Haematopoietic Stem cell (HSC) markers, suggesting an HSC mimicry, associated with higher ability to form mammospheres. Interestingly, N-CadherinHigh MDA cells showed greater capacity to adhere to SNOs, while the inhibition of SNO-mediating MDA cell proliferation was unremarkable. To investigate whether these features were shared by mouse BrCa, we used the 4T1 cell line in which N-Cadherin expression was abolished and then rescued. At variance with MDA cells, 4T1 cells expressing N-Cadherin revealed that the latter was associated with a lower expression of the HSC marker, Cxcr4, along with a lower capacity to form mammospheres. Furthermore, the rescue of N-Cadherin expression increased cell-cell adhesion and reduced proliferation of 4T1 cells when they were co-plated with SNOs. In conclusion, we demonstrated that: (i) N-CadherinHigh and Notch2High MDA cells showed similar HSC mimicry and dormancy features; (ii) N-Cadherin mediated BrCa-SNO adhesion; (iii) N-Cadherin had a positive Notch2-dependent role on SNO-induced dormancy and HSC mimicry in MDA cells, and a negative role in 4T1 cell stemness and HSC mimicry.
Keywords: BrCa dormancy; HSC-mimicry; bone microenvironment; cancer stem cells; endosteal niche; osteoblasts.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures




Similar articles
-
Notch2 pathway mediates breast cancer cellular dormancy and mobilisation in bone and contributes to haematopoietic stem cell mimicry.Br J Cancer. 2019 Jul;121(2):157-171. doi: 10.1038/s41416-019-0501-y. Epub 2019 Jun 26. Br J Cancer. 2019. PMID: 31239543 Free PMC article.
-
The bone metastasis niche in breast cancer-potential overlap with the haematopoietic stem cell niche in vivo.J Bone Oncol. 2019 Jun 7;17:100244. doi: 10.1016/j.jbo.2019.100244. eCollection 2019 Aug. J Bone Oncol. 2019. PMID: 31236323 Free PMC article.
-
E-cadherin expression in human breast cancer cells suppresses the development of osteolytic bone metastases in an experimental metastasis model.Cancer Res. 1996 Sep 1;56(17):4063-70. Cancer Res. 1996. PMID: 8752180
-
Regulation of stem cells in the niche.Cornea. 2005 Nov;24(8 Suppl):S12-S17. doi: 10.1097/01.ico.0000178742.98716.65. Cornea. 2005. PMID: 16227817 Review.
-
The stem cell niches in bone.J Clin Invest. 2006 May;116(5):1195-201. doi: 10.1172/JCI28568. J Clin Invest. 2006. PMID: 16670760 Free PMC article. Review.
Cited by
-
Bridging the Gap in Breast Cancer Dormancy: Models, Mechanisms, and Translational Challenges.Pharmaceuticals (Basel). 2025 Jun 26;18(7):961. doi: 10.3390/ph18070961. Pharmaceuticals (Basel). 2025. PMID: 40732251 Free PMC article. Review.
-
Osteoblasts and osteoclasts: an important switch of tumour cell dormancy during bone metastasis.J Exp Clin Cancer Res. 2022 Oct 28;41(1):316. doi: 10.1186/s13046-022-02520-0. J Exp Clin Cancer Res. 2022. PMID: 36307871 Free PMC article. Review.
-
The osteoblast in regulation of tumor cell dormancy and bone metastasis.J Bone Oncol. 2024 Mar 21;45:100597. doi: 10.1016/j.jbo.2024.100597. eCollection 2024 Apr. J Bone Oncol. 2024. PMID: 38550395 Free PMC article.
-
Muscle and Bone Defects in Metastatic Disease.Curr Osteoporos Rep. 2022 Oct;20(5):273-289. doi: 10.1007/s11914-022-00741-y. Epub 2022 Aug 22. Curr Osteoporos Rep. 2022. PMID: 35994202 Free PMC article. Review.
-
The Prognosis of Cancer Depends on the Interplay of Autophagy, Apoptosis, and Anoikis within the Tumor Microenvironment.Cell Biochem Biophys. 2023 Dec;81(4):621-658. doi: 10.1007/s12013-023-01179-4. Epub 2023 Oct 3. Cell Biochem Biophys. 2023. PMID: 37787970 Review.
References
-
- Howlader N., Noone A.M., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., et al. Cancer Statistics Review, 1975-2018-SEER Statistics. [(accessed on 4 October 2021)]; Available online: https://seer.cancer.gov/csr/1975_2018/
-
- Naumov G.N., Townson J.L., MacDonald I.C., Wilson S.M., Bramwell V.H.C., Groom A.C., Chambers A.F. Ineffectiveness of doxorubicin treatment on solitary dormant mammary carcinoma cells or late-developing metastases. Breast Cancer Res. Treat. 2003;82:199–206. doi: 10.1023/B:BREA.0000004377.12288.3c. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

