High-Throughput Monoclonal Antibody Discovery from Phage Libraries: Challenging the Current Preclinical Pipeline to Keep the Pace with the Increasing mAb Demand
- PMID: 35267633
- PMCID: PMC8909429
- DOI: 10.3390/cancers14051325
High-Throughput Monoclonal Antibody Discovery from Phage Libraries: Challenging the Current Preclinical Pipeline to Keep the Pace with the Increasing mAb Demand
Abstract
Monoclonal antibodies are among the most powerful therapeutics in modern medicine. Since the approval of the first therapeutic antibody in 1986, monoclonal antibodies keep holding great expectations for application in a range of clinical indications, highlighting the need to provide timely and sustainable access to powerful screening options. However, their application in the past has been limited by time-consuming and expensive steps of discovery and production. The screening of antibody repertoires is a laborious step; however, the implementation of next-generation sequencing-guided screening of single-chain antibody fragments has now largely overcome this issue. This review provides a detailed overview of the current strategies for the identification of monoclonal antibodies from phage display-based libraries. We also discuss the challenges and the possible solutions to improve the limiting selection and screening steps, in order to keep pace with the increasing demand for monoclonal antibodies.
Keywords: high-throughput screening; mAbs production; monoclonal antibodies; next-generation sequencing; selection-related; selection-unrelated; target-unrelated; third-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
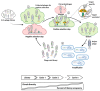


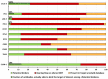



References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

