Cranberry Polyphenols in Esophageal Cancer Inhibition: New Insights
- PMID: 35267943
- PMCID: PMC8912450
- DOI: 10.3390/nu14050969
Cranberry Polyphenols in Esophageal Cancer Inhibition: New Insights
Abstract
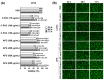
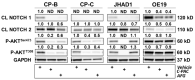
Esophageal adenocarcinoma (EAC) is a cancer characterized by rapidly rising incidence and poor survival, resulting in the need for new prevention and treatment options. We utilized two cranberry polyphenol extracts, one proanthocyanidin enriched (C-PAC) and a combination of anthocyanins, flavonoids, and glycosides (AFG) to assess inhibitory mechanisms utilizing premalignant Barrett's esophagus (BE) and EAC derived cell lines. We employed reverse phase protein arrays (RPPA) and Western blots to examine cancer-associated pathways and specific signaling cascades modulated by C-PAC or AFG. Viability results show that C-PAC is more potent than AFG at inducing cell death in BE and EAC cell lines. Based on the RPPA results, C-PAC significantly modulated 37 and 69 proteins in JH-EsoAd1 (JHAD1) and OE19 EAC cells, respectively. AFG treatment significantly altered 49 proteins in both JHAD1 and OE19 cells. Bioinformatic analysis of RPPA results revealed many previously unidentified pathways as modulated by cranberry polyphenols including NOTCH signaling, immune response, and epithelial to mesenchymal transition. Collectively, these results provide new insight regarding mechanisms by which cranberry polyphenols exert cancer inhibitory effects targeting EAC, with implications for potential use of cranberry constituents as cancer preventive agents.
Keywords: Barrett’s esophagus; anthocyanins; cranberry polyphenols; esophageal adenocarcinoma; flavonoids; glycosides; proanthocyanidins; reverse phase protein array.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Dulak A.M., Stojanov P., Peng S., Lawrence M.S., Fox C., Stewart C., Bandla S., Imamura Y., Schumacher S.E., Shefler E., et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013;45:478–486. doi: 10.1038/ng.2591. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

