Metabolite Dysregulation by Pranlukast in Mycobacterium tuberculosis
- PMID: 35268621
- PMCID: PMC8911922
- DOI: 10.3390/molecules27051520
Metabolite Dysregulation by Pranlukast in Mycobacterium tuberculosis
Abstract
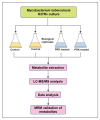
Mycobacterium tuberculosis has been infecting millions of people worldwide over the years, causing tuberculosis. Drugs targeting distinct cellular mechanisms including synthesis of the cell wall, lipids, proteins, and nucleic acids in Mtb are currently being used for the treatment of TB. Although extensive research is being carried out at the molecular level in the infected host and pathogen, the identification of suitable drug targets and drugs remains under explored. Pranlukast, an allosteric inhibitor of MtArgJ (Mtb ornithine acetyltransferase) has previously been shown to inhibit the survival and virulence of Mtb. The main objective of this study was to identify the altered metabolic pathways and biological processes associated with the differentially expressed metabolites by PRK in Mtb. Here in this study, metabolomics was carried out using an LC-MS/MS-based approach. Collectively, 50 metabolites were identified to be differentially expressed with a significant p-value through a global metabolomic approach using a high-resolution mass spectrometer. Metabolites downstream of argJ were downregulated in the arginine biosynthetic pathway following pranlukast treatment. Predicted human protein interactors of pranlukast-treated Mtb metabolome were identified in association with autophagy, inflammation, DNA repair, and other immune-related processes. Further metabolites including N-acetylglutamate, argininosuccinate, L-arginine, succinate, ergothioneine, and L-phenylalanine were validated by multiple reaction monitoring, a targeted mass spectrometry-based metabolomic approach. This study facilitates the understanding of pranlukast-mediated metabolic changes in Mtb and holds the potential to identify novel therapeutic approaches using metabolic pathways in Mtb.
Keywords: antagonist; bacteria; mass spectrometer; targeted metabolomics; untargeted metabolomics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Dookie N., Rambaran S., Padayatchi N., Mahomed S., Naidoo K. Evolution of drug resistance in Mycobacterium tuberculosis: A review on the molecular determinants of resistance and implications for personalized care. J. Antimicrob. Chemother. 2018;73:1138–1151. doi: 10.1093/jac/dkx506. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

