Optimization of Biocompatibility for a Hydrophilic Biological Molecule Encapsulation System
- PMID: 35268673
- PMCID: PMC8911823
- DOI: 10.3390/molecules27051572
Optimization of Biocompatibility for a Hydrophilic Biological Molecule Encapsulation System
Abstract
Despite considerable advances in recent years, challenges in delivery and storage of biological drugs persist and may delay or prohibit their clinical application. Though nanoparticle-based approaches for small molecule drug encapsulation are mature, encapsulation of proteins remains problematic due to destabilization of the protein. Reverse micelles composed of decylmonoacyl glycerol (10MAG) and lauryldimethylamino-N-oxide (LDAO) in low-viscosity alkanes have been shown to preserve the structure and stability of a wide range of biological macromolecules. Here, we present a first step on developing this system as a future platform for storage and delivery of biological drugs by replacing the non-biocompatible alkane solvent with solvents currently used in small molecule delivery systems. Using a novel screening approach, we performed a comprehensive evaluation of the 10MAG/LDAO system using two preparation methods across seven biocompatible solvents with analysis of toxicity and encapsulation efficiency for each solvent. By using an inexpensive hydrophilic small molecule to test a wide range of conditions, we identify optimal solvent properties for further development. We validate the predictions from this screen with preliminary protein encapsulation tests. The insight provided lays the foundation for further development of this system toward long-term room-temperature storage of biologics or toward water-in-oil-in-water biologic delivery systems.
Keywords: fluorescence spectroscopy; protein encapsulation; reverse micelle; viability.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
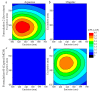
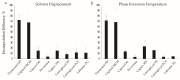
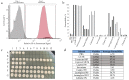
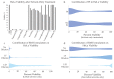

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

