Indole-Based Tubulin Inhibitors: Binding Modes and SARs Investigations
- PMID: 35268688
- PMCID: PMC8911766
- DOI: 10.3390/molecules27051587
Indole-Based Tubulin Inhibitors: Binding Modes and SARs Investigations
Abstract
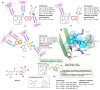
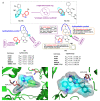
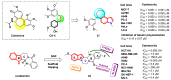
Tubulin inhibitors can interfere with normal cell mitosis and inhibit cell proliferation through interfering with the normal structure and function of microtubules, forming spindle filaments. Indole, as a privileged pharmacological skeleton, has been widely used in anti-cancer inhibitors. A variety of alkaloids containing an indole core obtained from natural sources have been proven to inhibit tubulin polymerization, and an ever-increasing number of synthetic indole-based tubulin inhibitors have been reported. Among these, several kinds of indole-based derivatives, such as TMP analogues, aroylindoles, arylthioindoles, fused indole, carbazoles, azacarbolines, alkaloid nortopsentin analogues and bis-indole derivatives, have shown good inhibition activities towards tubulin polymerization. The binding modes and SARs investigations of synthetic indole derivatives, along with a brief mechanism on their anti-tubulin activity, are presented in this review.
Keywords: SARs investigations; binding modes; cancer; indole; tubulin inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



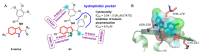










Similar articles
-
Indole molecules as inhibitors of tubulin polymerization: potential new anticancer agents.Future Med Chem. 2012 Oct;4(16):2085-115. doi: 10.4155/fmc.12.141. Future Med Chem. 2012. PMID: 23157240 Review.
-
Indole, a core nucleus for potent inhibitors of tubulin polymerization.Med Res Rev. 2007 Mar;27(2):209-38. doi: 10.1002/med.20080. Med Res Rev. 2007. PMID: 16788980 Review.
-
Synthesis and Preclinical Evaluation of Indole Triazole Conjugates as Microtubule Targeting Agents that are Effective against MCF-7 Breast Cancer Cell Lines.Anticancer Agents Med Chem. 2021;21(8):1047-1055. doi: 10.2174/1871520620666200925102940. Anticancer Agents Med Chem. 2021. PMID: 32981511
-
New 6- and 7-heterocyclyl-1H-indole derivatives as potent tubulin assembly and cancer cell growth inhibitors.Eur J Med Chem. 2018 May 25;152:283-297. doi: 10.1016/j.ejmech.2018.04.042. Epub 2018 Apr 25. Eur J Med Chem. 2018. PMID: 29730191
-
Design, synthesis and biological evaluation of 2-alkoxycarbonyl-3-anilinoindoles as a new class of potent inhibitors of tubulin polymerization.Bioorg Chem. 2020 Apr;97:103665. doi: 10.1016/j.bioorg.2020.103665. Epub 2020 Feb 18. Bioorg Chem. 2020. PMID: 32086053
Cited by
-
Targeting Histone Deacetylases 6 in Dual-Target Therapy of Cancer.Pharmaceutics. 2023 Nov 3;15(11):2581. doi: 10.3390/pharmaceutics15112581. Pharmaceutics. 2023. PMID: 38004560 Free PMC article. Review.
-
Novel Indole-Containing Hybrids Derived from Millepachine: Synthesis, Biological Evaluation and Antitumor Mechanism Study.Molecules. 2023 Feb 3;28(3):1481. doi: 10.3390/molecules28031481. Molecules. 2023. PMID: 36771147 Free PMC article.
-
Recent Advances of Tubulin Inhibitors Targeting the Colchicine Binding Site for Cancer Therapy.Biomolecules. 2022 Dec 10;12(12):1843. doi: 10.3390/biom12121843. Biomolecules. 2022. PMID: 36551271 Free PMC article. Review.
-
Evaluation of Antimicrobial, Anticholinesterase Potential of Indole Derivatives and Unexpectedly Synthesized Novel Benzodiazine: Characterization, DFT and Hirshfeld Charge Analysis.Molecules. 2023 Jun 27;28(13):5024. doi: 10.3390/molecules28135024. Molecules. 2023. PMID: 37446687 Free PMC article.
-
Design, Synthesis, and Antiproliferative Activity of Novel Indole/1,2,4-Triazole Hybrids as Tubulin Polymerization Inhibitors.Pharmaceuticals (Basel). 2025 Feb 19;18(2):275. doi: 10.3390/ph18020275. Pharmaceuticals (Basel). 2025. PMID: 40006087 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

