Molecular Dynamics Simulation Study of the Self-Assembly of Phenylalanine Peptide Nanotubes
- PMID: 35269349
- PMCID: PMC8912360
- DOI: 10.3390/nano12050861
Molecular Dynamics Simulation Study of the Self-Assembly of Phenylalanine Peptide Nanotubes
Abstract
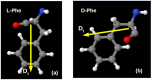
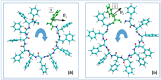
In this paper, we propose and use a new approach for a relatively simple technique for conducting MD simulation (MDS) of various molecular nanostructures, determining the trajectory of the MD run and forming the final structure using external force actions. A molecular dynamics manipulator (MD manipulator) is a controlled MDS type. As an example, the applicability of the developed algorithm for assembling peptide nanotubes (PNT) from linear phenylalanine (F or Phe) chains of different chirality is presented. The most adequate regimes for the formation of nanotubes of right chirality D from the initial L-F and nanotubes of left chirality L of their initial dipeptides D-F modes were determined. We use the method of a mixed (vector-scalar) product of the vectors of the sequence of dipole moments of phenylalanine molecules located along the nanotube helix to calculate the magnitude and sign of chirality of self-assembled helical phenylalanine nanotubes, which shows the validity of the proposed approach. As result, all data obtained correspond to the regularity of the chirality sign change of the molecular structures with a hierarchical complication of their organization.
Keywords: MD manipulator; chirality; controlled molecular dynamics; molecular dynamics method; nanotubes; phenylalanine; self-assembly of nanostructures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Orsi M. Molecular simulation of self-assembly. In: Azevedo H.S., da Silva R.M.P., editors. Self-Assembling Biomaterials. Molecular Design, Characterization and Application in Biology and Medicine. 1st ed. Elsevier; Amsterdam, The Netherlands: 2018. pp. 305–318. (Woodhead Publishing Series in Biomaterials).
-
- Van Gunsteren W.F., Berendsen H.J.C. Computer Simulation of Molecular Dynamics: Methodology, Applications, and Perspectives in Chemistry. Angew. Chem. Int. Ed. Engl. 1990;29:9921023. doi: 10.1002/anie.199009921. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

