CRISPR/Cas9-Mediated Models of Retinitis Pigmentosa Reveal Differential Proliferative Response of Müller Cells between Xenopus laevis and Xenopus tropicalis
- PMID: 35269429
- PMCID: PMC8909648
- DOI: 10.3390/cells11050807
CRISPR/Cas9-Mediated Models of Retinitis Pigmentosa Reveal Differential Proliferative Response of Müller Cells between Xenopus laevis and Xenopus tropicalis
Abstract
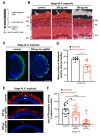
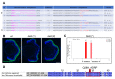
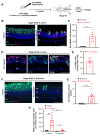
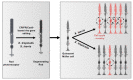
Retinitis pigmentosa is an inherited retinal dystrophy that ultimately leads to blindness due to the progressive degeneration of rod photoreceptors and the subsequent non-cell autonomous death of cones. Rhodopsin is the most frequently mutated gene in this disease. We here developed rhodopsin gene editing-based models of retinitis pigmentosa in two Xenopus species, Xenopus laevis and Xenopus tropicalis, by using CRISPR/Cas9 technology. In both of them, loss of rhodopsin function results in massive rod cell degeneration characterized by progressive shortening of outer segments and occasional cell death. This is followed by cone morphology deterioration. Despite these apparently similar degenerative environments, we found that Müller glial cells behave differently in Xenopus laevis and Xenopus tropicalis. While a significant proportion of Müller cells re-enter into the cell cycle in Xenopus laevis, their proliferation remains extremely limited in Xenopus tropicalis. This work thus reveals divergent responses to retinal injury in closely related species. These models should help in the future to deepen our understanding of the mechanisms that have shaped regeneration during evolution, with tremendous differences across vertebrates.
Keywords: CRISPR/Cas9; Müller cells; photoreceptors; retina; rhodopsin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

