The Cell Membrane of a Novel Rhizobium phaseoli Strain Is the Crucial Target for Aluminium Toxicity and Tolerance
- PMID: 35269493
- PMCID: PMC8909678
- DOI: 10.3390/cells11050873
The Cell Membrane of a Novel Rhizobium phaseoli Strain Is the Crucial Target for Aluminium Toxicity and Tolerance
Abstract
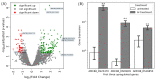
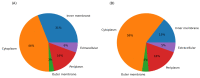
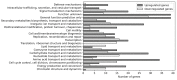
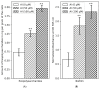
Soils with low pH and high aluminium (Al) contamination restrict common bean production, mainly due to adverse effects on rhizobia. We isolated a novel rhizobium strain, B3, from Kenyan soil which is more tolerant to Al stress than the widely used commercial strain CIAT899. B3 was resistant to 50 µM Al and recovered from 100 µM Al stress, while CIAT899 did not. Calcein labeling showed that less Al binds to the B3 membranes and less ATP and mScarlet-1 protein, a cytoplasmic marker, leaked out of B3 than CIAT899 cells in Al-containing media. Expression profiles showed that the primary targets of Al are genes involved in membrane biogenesis, metal ions binding and transport, carbohydrate, and amino acid metabolism and transport. The identified differentially expressed genes suggested that the intracellular γ-aminobutyric acid (GABA), glutathione (GSH), and amino acid levels, as well as the amount of the extracellular exopolysaccharide (EPS), might change during Al stress. Altered EPS levels could also influence biofilm formation. Therefore, these parameters were investigated in more detail. The GABA levels, extracellular EPS production, and biofilm formation increased, while GSH and amino acid level decreased. In conclusion, our comparative analysis identified genes that respond to Al stress in R. phaseoli. It appears that a large portion of the identified genes code for proteins stabilizing the plasma membrane. These genes might be helpful for future studies investigating the molecular basis of Al tolerance and the characterization of candidate rhizobial isolates that perform better in Al-contaminated soils than commercial strains.
Keywords: RNA-Seq; Rhizobium phaseoli; aluminium toxicity; aluminum tolerance; common bean; gene expression.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





Similar articles
-
Isolation and Characterization of High-Efficiency Rhizobia From Western Kenya Nodulating With Common Bean.Front Microbiol. 2021 Sep 10;12:697567. doi: 10.3389/fmicb.2021.697567. eCollection 2021. Front Microbiol. 2021. PMID: 34566909 Free PMC article.
-
Extracellular Polysaccharide Is Not Responsible for Aluminum Tolerance of Rhizobium leguminosarum bv. Phaseoli CIAT899.Appl Environ Microbiol. 1992 Apr;58(4):1095-101. doi: 10.1128/aem.58.4.1095-1101.1992. Appl Environ Microbiol. 1992. PMID: 16348680 Free PMC article.
-
Efficient nitrogen-fixing Rhizobium strains isolated from amazonian soils are highly tolerant to acidity and aluminium.World J Microbiol Biotechnol. 2012 May;28(5):1947-59. doi: 10.1007/s11274-011-0997-7. Epub 2012 Jan 6. World J Microbiol Biotechnol. 2012. PMID: 22806016
-
Nature and mechanisms of aluminium toxicity, tolerance and amelioration in symbiotic legumes and rhizobia.Biol Fertil Soils. 2018;54(3):309-318. doi: 10.1007/s00374-018-1262-0. Epub 2018 Feb 12. Biol Fertil Soils. 2018. PMID: 31258230 Free PMC article. Review.
-
Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants.Crit Rev Biotechnol. 2021 Aug;41(5):715-730. doi: 10.1080/07388551.2021.1874282. Epub 2021 Apr 18. Crit Rev Biotechnol. 2021. PMID: 33866893 Review.
Cited by
-
An Oxalate Transporter Gene, AtOT, Enhances Aluminum Tolerance in Arabidopsis thaliana by Regulating Oxalate Efflux.Int J Mol Sci. 2023 Feb 24;24(5):4516. doi: 10.3390/ijms24054516. Int J Mol Sci. 2023. PMID: 36901947 Free PMC article.
-
Distribution, Characterization and the Commercialization of Elite Rhizobia Strains in Africa.Int J Mol Sci. 2022 Jun 13;23(12):6599. doi: 10.3390/ijms23126599. Int J Mol Sci. 2022. PMID: 35743041 Free PMC article. Review.
-
Cloning, Expression Analysis, and Functional Characterization of Candidate Oxalate Transporter Genes of HbOT1 and HbOT2 from Rubber Tree (Hevea brasiliensis).Cells. 2022 Nov 27;11(23):3793. doi: 10.3390/cells11233793. Cells. 2022. PMID: 36497054 Free PMC article.
-
Rhizobia Contribute to Salinity Tolerance in Common Beans (Phaseolus vulgaris L.).Cells. 2022 Nov 16;11(22):3628. doi: 10.3390/cells11223628. Cells. 2022. PMID: 36429056 Free PMC article.
-
Evaluation of Legume-Rhizobial Symbiotic Interactions Beyond Nitrogen Fixation That Help the Host Survival and Diversification in Hostile Environments.Microorganisms. 2023 May 31;11(6):1454. doi: 10.3390/microorganisms11061454. Microorganisms. 2023. PMID: 37374957 Free PMC article. Review.
References
-
- FAOSTAT Food and Agricultural Organization of the United Nations. 2019. [(accessed on 15 December 2021)]. Available online: https://www.fao.org/faostat/en/#data.
-
- Herridge D.F., Peoples M.B., Boddey R.M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil. 2008;311:1–18. doi: 10.1007/s11104-008-9668-3. - DOI
-
- Sharma V., Bhattacharyya S., Kumar R., Kumar A., Ibañez F., Wang J., Guo B., Sudini H.K., Gopalakrishnan S., DasGupta M. Molecular basis of root nodule symbiosis between Bradyrhizobium and ‘crack-entry’legume groundnut (Arachis hypogaea L.) Plants. 2020;9:276. doi: 10.3390/plants9020276. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

