Production of an Active, Human Membrane Protein in Saccharomyces cerevisiae: Full-Length FICD
- PMID: 35269596
- PMCID: PMC8910494
- DOI: 10.3390/ijms23052458
Production of an Active, Human Membrane Protein in Saccharomyces cerevisiae: Full-Length FICD
Abstract
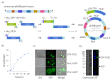
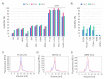
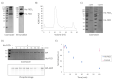
The human Fic domain-containing protein (FICD) is a type II endoplasmic reticulum (ER) membrane protein that is important for the maintenance of ER proteostasis. Structural and in vitro biochemical characterisation of FICD AMPylase and deAMPylase activity have been restricted to the soluble ER-luminal domain produced in Escherichia coli. Information about potentially important features, such as structural motifs, modulator binding sites or other regulatory elements, is therefore missing for the approximately 100 N-terminal residues including the transmembrane region of FICD. Expressing and purifying the required quantity and quality of membrane proteins is demanding because of the low yields and poor stability often observed. Here, we produce full-length FICD by combining a Saccharomyces cerevisiae-based platform with green fluorescent protein (GFP) tagging to optimise the conditions for expression, solubilisation and purification. We subsequently employ these conditions to purify milligram quantities of His-tagged FICD per litre of culture, and show that the purified, detergent-solubilised membrane protein is an active deAMPylating enzyme. Our work provides a straightforward methodology for producing not only full-length FICD, but also other membrane proteins in S. cerevisiae for structural and biochemical characterisation.
Keywords: AMPylation; FICD; Fic proteins; Saccharomyces cerevisiae; membrane protein purification; recombinant protein expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sanyal A., Chen A.J., Nakayasu E.S., Lazar C.S., Zbornik E.A., Worby C.A., Koller A., Mattoo S. A novel link between Fic (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J. Biol. Chem. 2015;290:8482–8499. doi: 10.1074/jbc.M114.618348. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

