Secondary Structure of Influenza A Virus Genomic Segment 8 RNA Folded in a Cellular Environment
- PMID: 35269600
- PMCID: PMC8910647
- DOI: 10.3390/ijms23052452
Secondary Structure of Influenza A Virus Genomic Segment 8 RNA Folded in a Cellular Environment
Abstract
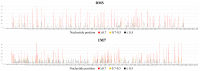
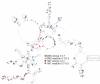
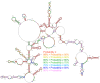
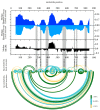
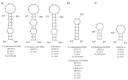
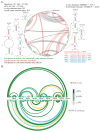
Influenza A virus (IAV) is a member of the single-stranded RNA (ssRNA) family of viruses. The most recent global pandemic caused by the SARS-CoV-2 virus has shown the major threat that RNA viruses can pose to humanity. In comparison, influenza has an even higher pandemic potential as a result of its high rate of mutations within its relatively short (<13 kbp) genome, as well as its capability to undergo genetic reassortment. In light of this threat, and the fact that RNA structure is connected to a broad range of known biological functions, deeper investigation of viral RNA (vRNA) structures is of high interest. Here, for the first time, we propose a secondary structure for segment 8 vRNA (vRNA8) of A/California/04/2009 (H1N1) formed in the presence of cellular and viral components. This structure shows similarities with prior in vitro experiments. Additionally, we determined the location of several well-defined, conserved structural motifs of vRNA8 within IAV strains with possible functionality. These RNA motifs appear to fold independently of regional nucleoprotein (NP)-binding affinity, but a low or uneven distribution of NP in each motif region is noted. This research also highlights several accessible sites for oligonucleotide tools and small molecules in vRNA8 in a cellular environment that might be a target for influenza A virus inhibition on the RNA level.
Keywords: IAV; RNA chemical mapping; RNA secondary structure; RNA virus; influenza A virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- World Health Organisation . Global Influenza Strategy Summary 2019–2030 Influenza. World Health Organisation; Geneva, Switzerland: 2019. pp. 1–2.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

