Production and Stabilization of Specific Upregulated Long Noncoding RNA HOXD-AS2 in Glioblastomas Are Mediated by TFE3 and miR-661, Respectively
- PMID: 35269968
- PMCID: PMC8911140
- DOI: 10.3390/ijms23052828
Production and Stabilization of Specific Upregulated Long Noncoding RNA HOXD-AS2 in Glioblastomas Are Mediated by TFE3 and miR-661, Respectively
Erratum in
-
Correction: Qin et al. Production and Stabilization of Specific Upregulated Long Noncoding RNA HOXD-AS2 in Glioblastomas Are Mediated by TFE3 and miR-661, Respectively. Int. J. Mol. Sci. 2022, 23, 2828.Int J Mol Sci. 2024 Dec 31;26(1):263. doi: 10.3390/ijms26010263. Int J Mol Sci. 2024. PMID: 39796287 Free PMC article.
Abstract
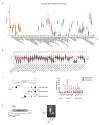
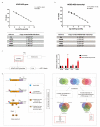
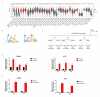
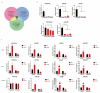
Differential expression of long noncoding RNAs (lncRNA) plays a key role in the development of gliomas. Because gliomas are the most common primary central nervous system tumor and glioblastomas have poor prognosis, it is urgent to develop new diagnostic methods. We have previously reported that lncRNA HOXD-AS2, which is specifically up-regulated in gliomas, can activate cell cycle and promote the development of gliomas. It is expected to be a new marker for molecular diagnosis of gliomas, but little is known about HOXD-AS2. Here, we demonstrate that TFE3 and miR-661 maintain the high expression level of HOXD-AS2 by regulating its production and degradation. We found that TFE3 acted as a transcription factor binding to the HOXD-AS2 promoter region and raised H3K27ac to activate HOXD-AS2. As the cytoplasmic-located lncRNA, HOXD-AS2 could be degraded by miR-661. This process was inhibited in gliomas due to the low expression of miR-661. Our study explains why HOXD-AS2 was specifically up-regulated in gliomas, helps to understand the molecular characteristics of gliomas, and provids insights for the search for specific markers in gliomas.
Keywords: HOXD-AS2; TFE3; glioma; miR-661.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Long non-coding RNA (lncRNA) HOXD-AS2 promotes glioblastoma cell proliferation, migration and invasion by regulating the miR-3681-5p/MALT1 signaling pathway.Bioengineered. 2021 Dec;12(2):9113-9127. doi: 10.1080/21655979.2021.1977104. Bioengineered. 2021. PMID: 34802389 Free PMC article.
-
Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop.J Exp Clin Cancer Res. 2020 Sep 22;39(1):196. doi: 10.1186/s13046-020-01695-8. J Exp Clin Cancer Res. 2020. PMID: 32962742 Free PMC article.
-
Promoter and enhancer RNAs regulate chromatin reorganization and activation of miR-10b/HOXD locus, and neoplastic transformation in glioma.Mol Cell. 2022 May 19;82(10):1894-1908.e5. doi: 10.1016/j.molcel.2022.03.018. Epub 2022 Apr 6. Mol Cell. 2022. PMID: 35390275 Free PMC article.
-
ADAMTS9-AS2: A Functional Long Non-coding RNA in Tumorigenesis.Curr Pharm Des. 2021;27(23):2722-2727. doi: 10.2174/1381612827666210325105106. Curr Pharm Des. 2021. PMID: 33823762 Review.
-
Long Non-coding RNA DLGAP1-AS1 and DLGAP1-AS2: Two Novel Oncogenes in Multiple Cancers.Curr Med Chem. 2023;30(25):2822-2834. doi: 10.2174/0929867329666220919114919. Curr Med Chem. 2023. PMID: 36121088 Review.
Cited by
-
HOXD-AS2-STAT3 feedback loop attenuates sensitivity to temozolomide in glioblastoma.CNS Neurosci Ther. 2023 Nov;29(11):3430-3445. doi: 10.1111/cns.14277. Epub 2023 Jun 12. CNS Neurosci Ther. 2023. PMID: 37308741 Free PMC article.
-
Computational Screening to Predict MicroRNA Targets in the Flavivirus 3' UTR Genome: An Approach for Antiviral Development.Int J Mol Sci. 2024 Sep 21;25(18):10135. doi: 10.3390/ijms251810135. Int J Mol Sci. 2024. PMID: 39337625 Free PMC article.
-
LncRNA HOXD-AS2 regulates miR-3681-5p/DCP1A axis to promote the progression of non-small cell lung cancer.J Thorac Dis. 2023 Mar 31;15(3):1289-1301. doi: 10.21037/jtd-23-153. Epub 2023 Mar 27. J Thorac Dis. 2023. PMID: 37065560 Free PMC article.
-
Correction: Qin et al. Production and Stabilization of Specific Upregulated Long Noncoding RNA HOXD-AS2 in Glioblastomas Are Mediated by TFE3 and miR-661, Respectively. Int. J. Mol. Sci. 2022, 23, 2828.Int J Mol Sci. 2024 Dec 31;26(1):263. doi: 10.3390/ijms26010263. Int J Mol Sci. 2024. PMID: 39796287 Free PMC article.
References
-
- Mizumoto M., Yamamoto T., Ishikawa E., Matsuda M., Takano S., Ishikawa H., Okumura T., Sakurai H., Matsumura A., Tsuboi K. Proton beam therapy with concurrent chemotherapy for glioblastoma multiforme: Comparison of nimustine hydrochloride and temozolomide. J. Neurooncol. 2016;130:165–170. doi: 10.1007/s11060-016-2228-4. - DOI - PubMed
-
- Eckel-Passow J.E., Lachance D.H., Molinaro A.M., Walsh K.M., Decker P.A., Sicotte H., Pekmezci M., Rice T., Kosel M.L., Smirnov I.V., et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

