Mutations of Omicron Variant at the Interface of the Receptor Domain Motif and Human Angiotensin-Converting Enzyme-2
- PMID: 35270013
- PMCID: PMC8911136
- DOI: 10.3390/ijms23052870
Mutations of Omicron Variant at the Interface of the Receptor Domain Motif and Human Angiotensin-Converting Enzyme-2
Abstract
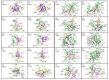
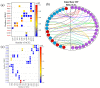
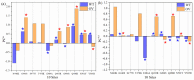
The most recent Omicron variant of SARS-CoV-2 has caused global concern and anxiety. The only thing certain about this strain, with a large number of mutations in the spike protein, is that it spreads quickly, seems to evade immune defense, and mitigates the benefits of existing vaccines. Based on the ultra-large-scale ab initio computational modeling of the receptor binding motif (RBM) and the human angiotensin-converting enzyme-2 (ACE2) interface, we provide the details of the effect of Omicron mutations at the fundamental atomic scale level. In-depth analysis anchored in the novel concept of amino acid-amino acid bond pair units (AABPU) indicates that mutations in the Omicron variant are connected with (i) significant changes in the shape and structure of AABPU components, together with (ii) significant increase in the positive partial charge, which facilitates the interaction with ACE2. We have identified changes in bonding due to mutations in the RBM. The calculated bond order, based on AABPU, reveals that the Omicron mutations increase the binding strength of RBM to ACE2. Our findings correlate with and are instrumental to explain the current observations and can contribute to the prediction of next potential new variant of concern.
Keywords: AABP unit; Omicron variant; SARS-CoV-2; complexity in shape and volume; partial charge; spike protein; virial transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Mohammad T., Shamsi A., Anwar S., Umair M., Hussain A., Rehman M.T., AlAjmi M.F., Islam A., Hassan M.I. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: Towards the development of effective COVID-19 therapy. Virus Res. 2020;288:198102. doi: 10.1016/j.virusres.2020.198102. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

