Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation
- PMID: 35271300
- PMCID: PMC9306331
- DOI: 10.1126/science.abn8933
Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation
Abstract
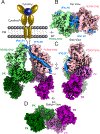
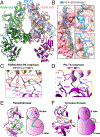
Cytokines signal through cell surface receptor dimers to initiate activation of intracellular Janus kinases (JAKs). We report the 3.6-angstrom-resolution cryo-electron microscopy structure of full-length JAK1 complexed with a cytokine receptor intracellular domain Box1 and Box2 regions captured as an activated homodimer bearing the valine→phenylalanine (VF) mutation prevalent in myeloproliferative neoplasms. The seven domains of JAK1 form an extended structural unit, the dimerization of which is mediated by close-packing of the pseudokinase (PK) domains from the monomeric subunits. The oncogenic VF mutation lies within the core of the JAK1 PK interdimer interface, enhancing packing complementarity to facilitate ligand-independent activation. The carboxy-terminal tyrosine kinase domains are poised for transactivation and to phosphorylate the receptor STAT (signal transducer and activator of transcription)-recruiting motifs projecting from the overhanging FERM (four-point-one, ezrin, radixin, moesin)-SH2 (Src homology 2)-domains. Mapping of constitutively active JAK mutants supports a two-step allosteric activation mechanism and reveals opportunities for selective therapeutic targeting of oncogenic JAK signaling.
Figures




Comment in
-
Unlocking the secrets to Janus kinase activation.Science. 2022 Apr 8;376(6589):139-140. doi: 10.1126/science.abo7788. Epub 2022 Apr 7. Science. 2022. PMID: 35389811
References
-
- Stroud RM, Wells JA, Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci STKE 2004, re7 (2004). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

