The changing epidemiology of SARS-CoV-2
- PMID: 35271324
- PMCID: PMC9009722
- DOI: 10.1126/science.abm4915
The changing epidemiology of SARS-CoV-2
Abstract
We have come a long way since the start of the COVID-19 pandemic-from hoarding toilet paper and wiping down groceries to sending our children back to school and vaccinating billions. Over this period, the global community of epidemiologists and evolutionary biologists has also come a long way in understanding the complex and changing dynamics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19. In this Review, we retrace our steps through the questions that this community faced as the pandemic unfolded. We focus on the key roles that mathematical modeling and quantitative analyses of empirical data have played in allowing us to address these questions and ultimately to better understand and control the pandemic.
Conflict of interest statement
Figures
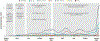
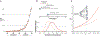

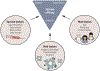
References
-
- Anderson RM, May RM, Infectious Diseases of Humans: Dynamics and Control (Oxford Univ. Press, 2010).
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

