HLA-DQ heterodimers in hematopoietic cell transplantation
- PMID: 35271697
- PMCID: PMC9121842
- DOI: 10.1182/blood.2022015860
HLA-DQ heterodimers in hematopoietic cell transplantation
Abstract
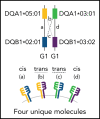
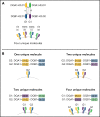
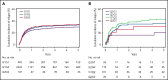
HLA-DQ heterodimers increase the susceptibility to autoimmune diseases, but their role in hematopoietic cell transplantation is unknown. We tested the hypothesis that outcome after HLA-matched and HLA-DQ-mismatched hematopoietic cell transplantation is influenced by HLA-DQ heterodimers. Heterodimers were defined in 5164 HLA-matched and 520 HLA-DQ-mismatched patients and their transplant donors according to well-established crystallographic criteria. Group 1 (G1) heterodimers are any DQA1*02/03/04/05/06α paired with any DQB1*02/03/04β. Group 2 (G2) heterodimers are DQA1*01α paired with any DQB1*05/06β. Multivariable models identified significantly higher relapse risk in G1G2 and G2G2 compared with G1G1 HLA-matched patients with malignant disease; risk increased with an increasing number of G2 molecules. In HLA-DQ-mismatched transplantation for malignant diseases, matching or mismatching for G2 increased relapse risk. G2 lowered disease-free survival after both HLA-matched and HLA-DQ-mismatched transplantation. A paradigm based on HLA-DQ heterodimers provides a functional definition of the hematopoietic cell transplantation barrier and a means to lower risks for future patients.
© 2022 by The American Society of Hematology.
Figures

Comment in
-
HLA-DQ + CD4 help = graft-versus-tumor effect.Blood. 2022 May 19;139(20):2999-3000. doi: 10.1182/blood.2022016117. Blood. 2022. PMID: 35587871 No abstract available.
References
-
- van Rood JJ, van Leeuwen A, Keuning JJ, van Oud Alblas AB. The serological recognition of the human MLC determinants using a modified cytotoxicity technique. Tissue Antigens. 1975;5(2):73-79. - PubMed
-
- Bodmer JG. Ia Serology. In: Bodmer WF, Batchelor JR, Bodmer, JG, eds. Histocompatibility Testing 1977. Copenhagen, Denmark: Munksgaard, 1978:351-357.
-
- Duquesnoy RJ, Marrari M, Annen K. Identification of an HLA-DR-associated system of B-cell alloantigens. Transplant Proc. 1979;11(4):1757-1760. - PubMed
-
- Terasaki PI, Park MS, Bernoco D, et al. Overview of the 1980 International Histocompatibility Workshop. In: Terasaki PI, ed. Histocompatibility Testing 1980. Los Angeles, CA: UCLA Tissue Typing Laboratory, 1980.
-
- McDevitt HO, Bodmer WF. HL-A, immune-response genes, and disease. Lancet. 1974;303(7869):1269-1275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

