IRAK4 Signaling Drives Resistance to Checkpoint Immunotherapy in Pancreatic Ductal Adenocarcinoma
- PMID: 35271824
- PMCID: PMC9387774
- DOI: 10.1053/j.gastro.2022.02.035
IRAK4 Signaling Drives Resistance to Checkpoint Immunotherapy in Pancreatic Ductal Adenocarcinoma
Abstract
Background & aims: Checkpoint immunotherapy is largely ineffective in pancreatic ductal adenocarcinoma (PDAC). The innate immune nuclear factor (NF)-κB pathway promotes PDAC cell survival and stromal fibrosis, and is driven by Interleukin-1 Receptor Associated Kinase-4 (IRAK4), but its impact on tumor immunity has not been directly investigated.
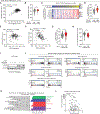
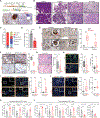
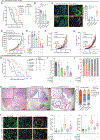
Methods: We interrogated The Cancer Genome Atlas data to identify the correlation between NF-κB and T cell signature, and a PDAC tissue microarray (TMA) to correlate IRAK4 activity with CD8+ T cell abundance. We performed RNA sequencing (RNA-seq) on IRAK4-deleted PDAC cells, and single-cell RNA-seq on autochthonous KPC (p48-Cre/TP53f/f/LSL-KRASG12D) mice treated with an IRAK4 inhibitor. We generated conditional IRAK4-deleted KPC mice and complementarily used IRAK4 inhibitors to determine the impact of IRAK4 on T cell immunity.
Results: We found positive correlation between NF-κB activity, IRAK4 and T cell exhaustion from The Cancer Genome Atlas. We observed inverse correlation between phosphorylated IRAK4 and CD8+ T cell abundance in a PDAC tissue microarray. Loss of IRAK4 abrogates NF-κB activity, several immunosuppressive factors, checkpoint ligands, and hyaluronan synthase 2, all of which drive T cell dysfunction. Accordingly, conditional deletion or pharmacologic inhibition of IRAK4 markedly decreased tumor desmoplasia and increased the abundance and activity of infiltrative CD4+ and CD8+ T cells in KPC tumors. Single-cell RNA-seq showed myeloid and fibroblast reprogramming toward acute inflammatory responses following IRAK4 inhibition. These changes set the stage for successful combination of IRAK4 inhibitors with checkpoint immunotherapy, resulting in excellent tumor control and markedly prolonged survival of KPC mice.
Conclusion: IRAK4 drives T cell dysfunction in PDAC and is a novel, promising immunotherapeutic target.
Keywords: CA-4948; HAS2; NF-κB; PD-L1; T cell exhaustion.
Copyright © 2022 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
the authors have declared no conflict of interests.
Figures




Comment in
-
Targeting IRAK4 Signaling in PDAC: Turning the "Cold" Tumors to "Hot" Ones.Gastroenterology. 2022 Jun;162(7):1837-1839. doi: 10.1053/j.gastro.2022.03.036. Epub 2022 Mar 28. Gastroenterology. 2022. PMID: 35358509 No abstract available.
References
-
- Dunne A, O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE 2003;2003:re3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

