An integrative gene expression signature analysis identifies CMS4 KRAS-mutated colorectal cancers sensitive to combined MEK and SRC targeted therapy
- PMID: 35272617
- PMCID: PMC8908604
- DOI: 10.1186/s12885-022-09344-3
An integrative gene expression signature analysis identifies CMS4 KRAS-mutated colorectal cancers sensitive to combined MEK and SRC targeted therapy
Abstract
Background: Over half of colorectal cancers (CRCs) are hard-wired to RAS/RAF/MEK/ERK pathway oncogenic signaling. However, the promise of targeted therapeutic inhibitors, has been tempered by disappointing clinical activity, likely due to complex resistance mechanisms that are not well understood. This study aims to investigate MEK inhibitor-associated resistance signaling and identify subpopulation(s) of CRC patients who may be sensitive to biomarker-driven drug combination(s).
Methods: We classified 2250 primary and metastatic human CRC tumors by consensus molecular subtypes (CMS). For each tumor, we generated multiple gene expression signature scores measuring MEK pathway activation, MEKi "bypass" resistance, SRC activation, dasatinib sensitivity, EMT, PC1, Hu-Lgr5-ISC, Hu-EphB2-ISC, Hu-Late TA, Hu-Proliferation, and WNT activity. We carried out correlation, survival and other bioinformatic analyses. Validation analyses were performed in two independent publicly available CRC tumor datasets (n = 585 and n = 677) and a CRC cell line dataset (n = 154).
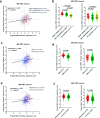
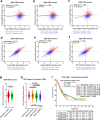
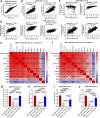
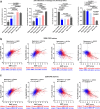
Results: Here we report a central role of SRC in mediating "bypass"-resistance to MEK inhibition (MEKi), primarily in cancer stem cells (CSCs). Our integrated and comprehensive gene expression signature analyses in 2250 CRC tumors reveal that MEKi-resistance is strikingly-correlated with SRC activation (Spearman P < 10-320), which is similarly associated with EMT (epithelial to mesenchymal transition), regional metastasis and disease recurrence with poor prognosis. Deeper analysis shows that both MEKi-resistance and SRC activation are preferentially associated with a mesenchymal CSC phenotype. This association is validated in additional independent CRC tumor and cell lines datasets. The CMS classification analysis demonstrates the strikingly-distinct associations of CMS1-4 subtypes with the MEKi-resistance and SRC activation. Importantly, MEKi + SRCi sensitivities are predicted to occur predominantly in the KRAS mutant, mesenchymal CSC-like CMS4 CRCs.
Conclusions: Large human tumor gene expression datasets representing CRC heterogeneity can provide deep biological insights heretofore not possible with cell line models, suggesting novel repurposed drug combinations. We identified SRC as a common targetable node--an Achilles' heel--in MEKi-targeted therapy-associated resistance in mesenchymal stem-like CRCs, which may help development of a biomarker-driven drug combination (MEKi + SRCi) to treat problematic subpopulations of CRC.
Keywords: CMS; Cancer stem cell; Colorectal cancer; EMT; Gene expression signature; MEK inhibitor; SRC; Targeted therapy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Ras Pathway Activation and MEKi Resistance Scores Predict the Efficiency of MEKi and SRCi Combination to Induce Apoptosis in Colorectal Cancer.Cancers (Basel). 2022 Mar 11;14(6):1451. doi: 10.3390/cancers14061451. Cancers (Basel). 2022. PMID: 35326598 Free PMC article.
-
Triple blockade of EGFR, MEK and PD-L1 has antitumor activity in colorectal cancer models with constitutive activation of MAPK signaling and PD-L1 overexpression.J Exp Clin Cancer Res. 2019 Dec 16;38(1):492. doi: 10.1186/s13046-019-1497-0. J Exp Clin Cancer Res. 2019. PMID: 31842958 Free PMC article.
-
Oncogenic KRAS-associated gene signature defines co-targeting of CDK4/6 and MEK as a viable therapeutic strategy in colorectal cancer.Oncogene. 2017 Aug 31;36(35):4975-4986. doi: 10.1038/onc.2017.120. Epub 2017 May 1. Oncogene. 2017. PMID: 28459468
-
Dual Inhibition of MEK and PI3K Pathway in KRAS and BRAF Mutated Colorectal Cancers.Int J Mol Sci. 2015 Sep 23;16(9):22976-88. doi: 10.3390/ijms160922976. Int J Mol Sci. 2015. PMID: 26404261 Free PMC article. Review.
-
MEK in cancer and cancer therapy.Pharmacol Ther. 2014 Feb;141(2):160-71. doi: 10.1016/j.pharmthera.2013.10.001. Epub 2013 Oct 9. Pharmacol Ther. 2014. PMID: 24121058 Review.
Cited by
-
Propofol enhances stem-like properties of glioma via GABAAR-dependent Src modulation of ZDHHC5-EZH2 palmitoylation mechanism.Stem Cell Res Ther. 2022 Aug 4;13(1):398. doi: 10.1186/s13287-022-03087-5. Stem Cell Res Ther. 2022. PMID: 35927718 Free PMC article.
-
Clinical Challenges of Consensus Molecular Subtype CMS4 Colon Cancer in the Era of Precision Medicine.Clin Cancer Res. 2024 Jun 3;30(11):2351-2358. doi: 10.1158/1078-0432.CCR-23-3964. Clin Cancer Res. 2024. PMID: 38564259 Free PMC article. Review.
-
An integrated model for predicting KRAS dependency.PLoS Comput Biol. 2023 May 4;19(5):e1011095. doi: 10.1371/journal.pcbi.1011095. eCollection 2023 May. PLoS Comput Biol. 2023. PMID: 37141389 Free PMC article.
-
Autofluorescent Cancer Stem Cells: Potential Biomarker to Predict Recurrence in Resected Colorectal Tumors.Cancer Res Commun. 2024 Oct 1;4(10):2575-2588. doi: 10.1158/2767-9764.CRC-24-0188. Cancer Res Commun. 2024. PMID: 39225547 Free PMC article.
-
Identifying distinct prognostic and predictive contributions of tumor epithelium versus tumor microenvironment in colorectal cancer.BMC Cancer. 2025 Mar 12;25(1):441. doi: 10.1186/s12885-025-13829-2. BMC Cancer. 2025. PMID: 40075322 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

