Nucleolar-based Dux repression is essential for embryonic two-cell stage exit
- PMID: 35273077
- PMCID: PMC8973846
- DOI: 10.1101/gad.349172.121
Nucleolar-based Dux repression is essential for embryonic two-cell stage exit
Abstract
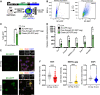
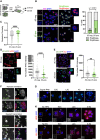
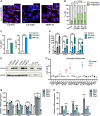
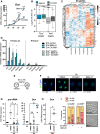
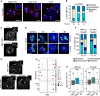
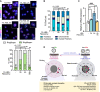
Upon fertilization, the mammalian embryo must switch from dependence on maternal transcripts to transcribing its own genome, and in mice this involves the transient up-regulation of MERVL transposons and MERVL-driven genes at the two-cell stage. The mechanisms and requirement for MERVL and two-cell (2C) gene up-regulation are poorly understood. Moreover, this MERVL-driven transcriptional program must be rapidly shut off to allow two-cell exit and developmental progression. Here, we report that robust ribosomal RNA (rRNA) synthesis and nucleolar maturation are essential for exit from the 2C state. 2C-like cells and two-cell embryos show similar immature nucleoli with altered structure and reduced rRNA output. We reveal that nucleolar disruption via blocking RNA polymerase I activity or preventing nucleolar phase separation enhances conversion to a 2C-like state in embryonic stem cells (ESCs) by detachment of the MERVL activator Dux from the nucleolar surface. In embryos, nucleolar disruption prevents proper nucleolar maturation and Dux silencing and leads to two- to four-cell arrest. Our findings reveal an intriguing link between rRNA synthesis, nucleolar maturation, and gene repression during early development.
Keywords: 2C-like state; Dux; MERVL; nucleolus; totipotency.
© 2022 Xie et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Azkanaz M, Rodríguez López A, de Boer B, Huiting W, Angrand PO, Vellenga E, Kampinga HH, Bergink S, Martens JH, Schuringa JJ, et al. 2019. Protein quality control in the nucleolus safeguards recovery of epigenetic regulators after heat shock. Elife 8: e45205. 10.7554/eLife.45205 - DOI - PMC - PubMed
-
- Binek A, Rojo D, Godzien J, Ruperez FJ, Nunez V, Jorge I, Ricote M, Vazquez J, Barbas C. 2019. Flow cytometry Has a significant impact on the cellular metabolome. J Proteome Res 18: 169–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
