Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis
- PMID: 35273176
- PMCID: PMC8913623
- DOI: 10.1038/s41467-022-28913-5
Fusobacterium nucleatum reduces METTL3-mediated m6A modification and contributes to colorectal cancer metastasis
Abstract
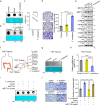
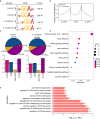
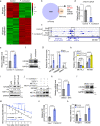
Microbiota-host interactions play critical roles in colorectal cancer (CRC) progression, however, the underlying mechanisms remain elusive. Here, we uncover that Fusobacterium nucleatum (F. nucleatum) induces a dramatic decline of m6A modifications in CRC cells and patient-derived xenograft (PDX) tissues by downregulation of an m6A methyltransferase METTL3, contributing to inducation of CRC aggressiveness. Mechanistically, we characterized forkhead box D3 (FOXD3) as a transcription factor for METTL3. F. nucleatum activates YAP signaling, inhibits FOXD3 expression, and subsequently reduces METTL3 transcription. Downregulation of METTL3 promotes its target kinesin family member 26B (KIF26B) expression by reducing its m6A levels and diminishing YTHDF2-dependent mRNA degradation, which contributes to F. nucleatum-induced CRC metastasis. Moreover, METTL3 expression is negatively correlated with F. nucleatum and KIF26B levels in CRC tissues. A high expression of KIF26B is also significantly correlated with a shorter survival time of CRC patients. Together, our findings provide insights into modulating human m6A epitranscriptome by gut microbiota, and its significance in CRC progression.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

