The SARS-CoV-2 Alpha variant exhibits comparable fitness to the D614G strain in a Syrian hamster model
- PMID: 35273335
- PMCID: PMC8913834
- DOI: 10.1038/s42003-022-03171-9
The SARS-CoV-2 Alpha variant exhibits comparable fitness to the D614G strain in a Syrian hamster model
Abstract
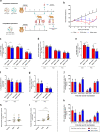
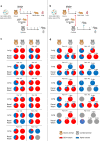
Late 2020, SARS-CoV-2 Alpha variant emerged in United Kingdom and gradually replaced G614 strains initially involved in the global spread of the pandemic. In this study, we use a Syrian hamster model to compare a clinical strain of Alpha variant with an ancestral G614 strain. The Alpha variant succeed to infect animals and to induce a pathology that mimics COVID-19. However, both strains replicate to almost the same level and induced a comparable disease and immune response. A slight fitness advantage is noted for the G614 strain during competition and transmission experiments. These data do not corroborate the epidemiological situation observed during the first half of 2021 in humans nor reports that showed a more rapid replication of Alpha variant in human reconstituted bronchial epithelium. This study highlights the need to combine data from different laboratories using various animal models to decipher the biological properties of newly emerging SARS-CoV-2 variants.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

