Atomic force microscopy-single-molecule force spectroscopy unveils GPCR cell surface architecture
- PMID: 35273337
- PMCID: PMC8913689
- DOI: 10.1038/s42003-022-03162-w
Atomic force microscopy-single-molecule force spectroscopy unveils GPCR cell surface architecture
Abstract
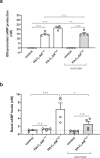
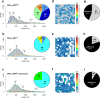
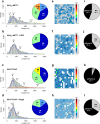
G protein-coupled receptors (GPCRs) form the largest family of cell surface receptors. Despite considerable insights into their pharmacology, the GPCR architecture at the cell surface still remains largely unexplored. Herein, we present the specific unfolding of different GPCRs at the surface of living mammalian cells by atomic force microscopy-based single molecule force spectroscopy (AFM-SMFS). Mathematical analysis of the GPCR unfolding distances at resting state revealed the presence of different receptor populations relying on distinct oligomeric states which are receptor-specific and receptor expression-dependent. Moreover, we show that the oligomer size dictates the receptor spatial organization with nanoclusters of high-order oligomers while lower-order complexes spread over the whole cell surface. Finally, the receptor activity reshapes both the oligomeric populations and their spatial arrangement. These results add an additional level of complexity to the GPCR pharmacology until now considered to arise from a single receptor population at the cell surface.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

