Complete Genome Sequence Analysis of Ralstonia solanacearum Strain PeaFJ1 Provides Insights Into Its Strong Virulence in Peanut Plants
- PMID: 35273586
- PMCID: PMC8904134
- DOI: 10.3389/fmicb.2022.830900
Complete Genome Sequence Analysis of Ralstonia solanacearum Strain PeaFJ1 Provides Insights Into Its Strong Virulence in Peanut Plants
Abstract
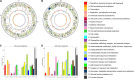
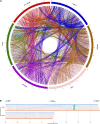
The bacterial wilt of peanut (Arachis hypogaea L.) caused by Ralstonia solanacearum is a devastating soil-borne disease that seriously restricted the world peanut production. However, the molecular mechanism of R. solanacearum-peanut interaction remains largely unknown. We found that R. solanacearum HA4-1 and PeaFJ1 isolated from peanut plants showed different pathogenicity by inoculating more than 110 cultivated peanuts. Phylogenetic tree analysis demonstrated that HA4-1 and PeaFJ1 both belonged to phylotype I and sequevar 14M, which indicates a high degree of genomic homology between them. Genomic sequencing and comparative genomic analysis of PeaFJ1 revealed 153 strain-specific genes compared with HA4-1. The PeaFJ1 strain-specific genes consisted of diverse virulence-related genes including LysR-type transcriptional regulators, two-component system-related genes, and genes contributing to motility and adhesion. In addition, the repertoire of the type III effectors of PeaFJ1 was bioinformatically compared with that of HA4-1 to find the candidate effectors responsible for their different virulences. There are 79 effectors in the PeaFJ1 genome, only 4 of which are different effectors compared with HA4-1, including RipS4, RipBB, RipBS, and RS_T3E_Hyp6. Based on the virulence profiles of the two strains against peanuts, we speculated that RipS4 and RipBB are candidate virulence effectors in PeaFJ1 while RipBS and RS_T3E_Hyp6 are avirulence effectors in HA4-1. In general, our research greatly reduced the scope of virulence-related genes and made it easier to find out the candidates that caused the difference in pathogenicity between the two strains. These results will help to reveal the molecular mechanism of peanut-R. solanacearum interaction and develop targeted control strategies in the future.
Keywords: Ralstonia solanacearum; comparative genomic analysis; genome sequencing; pathogenicity; peanut.
Copyright © 2022 Tan, Dai, Chen, Wu, Yang, Zheng, Chen, Wan and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Complete Genome Sequence of Sequevar 14M Ralstonia solanacearum Strain HA4-1 Reveals Novel Type III Effectors Acquired Through Horizontal Gene Transfer.Front Microbiol. 2019 Aug 14;10:1893. doi: 10.3389/fmicb.2019.01893. eCollection 2019. Front Microbiol. 2019. PMID: 31474968 Free PMC article.
-
Complete genome sequence analysis of the peanut pathogen Ralstonia solanacearum strain Rs-P.362200.BMC Microbiol. 2021 Apr 19;21(1):118. doi: 10.1186/s12866-021-02157-7. BMC Microbiol. 2021. PMID: 33874906 Free PMC article.
-
Comparative genomic analysis of Ralstonia solanacearum reveals candidate avirulence effectors in HA4-1 triggering wild potato immunity.Front Plant Sci. 2023 Feb 23;14:1075042. doi: 10.3389/fpls.2023.1075042. eCollection 2023. Front Plant Sci. 2023. PMID: 36909411 Free PMC article.
-
Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era.Mol Plant Pathol. 2013 Sep;14(7):651-62. doi: 10.1111/mpp.12038. Epub 2013 May 30. Mol Plant Pathol. 2013. PMID: 23718203 Free PMC article. Review.
-
Lessons learned from the genome analysis of ralstonia solanacearum.Annu Rev Phytopathol. 2004;42:107-34. doi: 10.1146/annurev.phyto.42.011204.104301. Annu Rev Phytopathol. 2004. PMID: 15283662 Review.
Cited by
-
Genome-wide analysis of the peanut CaM/CML gene family reveals that the AhCML69 gene is associated with resistance to Ralstonia solanacearum.BMC Genomics. 2024 Feb 21;25(1):200. doi: 10.1186/s12864-024-10108-5. BMC Genomics. 2024. PMID: 38378471 Free PMC article.
-
Transcriptome analysis reveals differential transcription in tomato (Solanum lycopersicum) following inoculation with Ralstonia solanacearum.Sci Rep. 2022 Dec 22;12(1):22137. doi: 10.1038/s41598-022-26693-y. Sci Rep. 2022. PMID: 36550145 Free PMC article.
-
Roles of AGD2a in Plant Development and Microbial Interactions of Lotus japonicus.Int J Mol Sci. 2022 Jun 20;23(12):6863. doi: 10.3390/ijms23126863. Int J Mol Sci. 2022. PMID: 35743304 Free PMC article.
-
Comparative transcriptome analysis revealed molecular mechanisms of peanut leaves responding to Ralstonia solanacearum and its type III secretion system mutant.Front Microbiol. 2022 Aug 25;13:998817. doi: 10.3389/fmicb.2022.998817. eCollection 2022. Front Microbiol. 2022. PMID: 36090119 Free PMC article.
-
Comprehensive genome sequence analysis of Ralstonia solanacearum gd-2, a phylotype I sequevar 15 strain collected from a tobacco bacterial phytopathogen.Front Microbiol. 2024 Mar 14;15:1335081. doi: 10.3389/fmicb.2024.1335081. eCollection 2024. Front Microbiol. 2024. PMID: 38550868 Free PMC article.
References
-
- Angot A., Peeters N., Lechner E., Vailleau F., Baud C., Gentzbittel L., et al. (2006). Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U.S.A. 103 14620–14625. 10.1073/pnas.0509393103 - DOI - PMC - PubMed
-
- Attieh Z., Mouawad C., Rejasse A., Jehanno I., Perchat S., I, Hegna K., et al. (2020). The fliK Gene Is Required for the Resistance of Bacillus thuringiensis to Antimicrobial Peptides and Virulence in Drosophila melanogaster. Front. Microbiol. 11:611220. 10.3389/fmicb.2020.611220 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

