Dominant CD8+ T Cell Nucleocapsid Targeting in SARS-CoV-2 Infection and Broad Spike Targeting From Vaccination
- PMID: 35273611
- PMCID: PMC8902813
- DOI: 10.3389/fimmu.2022.835830
Dominant CD8+ T Cell Nucleocapsid Targeting in SARS-CoV-2 Infection and Broad Spike Targeting From Vaccination
Abstract
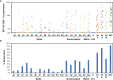
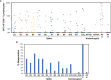
CD8+ T cells have key protective roles in many viral infections. While an overall Th1-biased cellular immune response against SARS-CoV-2 has been demonstrated, most reports of anti-SARS-CoV-2 cellular immunity have evaluated bulk T cells using pools of predicted epitopes, without clear delineation of the CD8+ subset and its magnitude and targeting. In recently infected persons (mean 29.8 days after COVID-19 symptom onset), we confirm a Th1 bias (and a novel IL-4-producing population of unclear significance) by flow cytometry, which does not correlate to antibody responses against the receptor binding domain. Evaluating isolated CD8+ T cells in more detail by IFN-γ ELISpot assays, responses against spike, nucleocapsid, matrix, and envelope proteins average 396, 901, 296, and 0 spot-forming cells (SFC) per million, targeting 1.4, 1.5, 0.59, and 0.0 epitope regions respectively. Nucleocapsid targeting is dominant in terms of magnitude, breadth, and density of targeting. The magnitude of responses drops rapidly post-infection; nucleocapsid targeting is most sustained, and vaccination selectively boosts spike targeting. In SARS-CoV-2-naïve persons, evaluation of the anti-spike CD8+ T cell response soon after vaccination (mean 11.3 days) yields anti-spike CD8+ T cell responses averaging 2,463 SFC/million against 4.2 epitope regions, and targeting mirrors that seen in infected persons. These findings provide greater clarity on CD8+ T cell anti-SARS-CoV-2 targeting, breadth, and persistence, suggesting that nucleocapsid inclusion in vaccines could broaden coverage and durability.
Keywords: CD8+ T cells; COVID-19; COVID-19 vaccine; SARS-CoV-2; cellular immunity.
Copyright © 2022 Taus, Hofmann, Ibarrondo, Hausner, Fulcher, Krogstad, Ferbas, Tobin, Rimoin, Aldrovandi and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- O'brien MP, Forleo-Neto E, Musser BJ, Isa F, Chan KC, Sarkar N, et al. Subcutaneous REGEN-COV Antibody Combination for Covid-19 Prevention. medRxiv (2021). doi: 10.1101/2021.06.14.21258567 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

