Let-7c increases BACE2 expression by RNAa and decreases Aβ production
- PMID: 35273693
- PMCID: PMC8902526
Let-7c increases BACE2 expression by RNAa and decreases Aβ production
Abstract
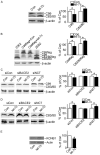
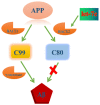
MicroRNAs (miRNAs) are highly conserved, non-coding transcripts that regulate gene expression in various ways. Evidence suggests that miRNAs may be a contributory factor in neurodegeneration, including Alzheimer's disease (AD), Parkinson's disease (PD), and triplet repeat disorders. In order to further understand the potential roles of miRNAs in the pathogenesis of AD, we analyzed Down syndrome (DS), a special model of AD, by using a TaqMan microRNA array and found that miRNA let-7c was up-regulated in both DS and AD. ELISA assay showed that let-7c reduced the expression level of Aβ significantly. Real-time quantitative-polymerase chain reaction (RT-qPCR) was conducted to reveal that the expression level of let-7c increased dramatically in DS cells, patients with DS and mice with AD compared with normal ones respectively. Additionally, western blotting illustrated that let-7c suppressed the expression of Aβ by inducing BACE2 to cut C99 and increase the content of C83/80. BACE2 expression was inhibited by let-7c and luciferase reporter gene assay revealed that let-7c increased the activity of wild-type BACE2 promoter but not 3'UTR. Furthermore, promoter analysis of BACE2 confirmed that let-7c could bind to BACE2 in the sequence between -1368 and -1347. In addition, immunoblotting assay demonstrated that let-7c induced BACE2 expression by RNAa. To the best of our knowledge, our study revealed for the first time that let-7c up-regulated BACE2 expression and decreased Aβ production.
Keywords: Alzheimer’s disease; BACE2; Let-7c; RNA activation; β-amyloid.
AJTR Copyright © 2022.
Conflict of interest statement
None.
Figures




References
-
- Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, Ledermann B, Burki K, Frey P, Paganetti PA, Waridel C, Calhoun ME, Jucker M, Probst A, Staufenbiel M, Sommer B. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc Natl Acad Sci U S A. 1997;94:13287–13292. - PMC - PubMed
-
- Kuo YM, Kokjohn TA, Beach TG, Sue LI, Brune D, Lopez JC, Kalback WM, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, Roher AE. Comparative analysis of amyloid-beta chemical structure and amyloid plaque morphology of transgenic mouse and Alzheimer’s disease brains. J Biol Chem. 2001;276:12991–12998. - PubMed
-
- Mori H, Takio K, Ogawara M, Selkoe DJ. Mass spectrometry of purified amyloid beta protein in Alzheimer’s disease. J Biol Chem. 1992;267:17082–17086. - PubMed
LinkOut - more resources
Full Text Sources
