The Gut Microbiota in Inflammatory Bowel Disease
- PMID: 35273921
- PMCID: PMC8902753
- DOI: 10.3389/fcimb.2022.733992
The Gut Microbiota in Inflammatory Bowel Disease
Abstract
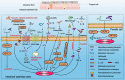
Epidemiological surveys indicate that the incidence of inflammatory bowel disease (IBD) is increasing rapidly with the continuous growth of the economy. A large number of studies have investigated the relationship between the genetic factors related to the susceptibility to IBD and the gut microbiota of patients by using high-throughput sequencing. IBD is considered the outcome of the interaction between host and microorganisms, including intestinal microbial factors, abnormal immune response, and a damaged intestinal mucosal barrier. The imbalance of microbial homeostasis leads to the colonization and invasion of opportunistic pathogens in the gut, which increases the risk of the host immune response and promotes the development of IBD. It is critical to identify the specific pathogens related to the pathogenesis of IBD. An in-depth understanding of various pathogenic factors is of great significance for the early detection of IBD. This review highlights the role of gut microbiota in the pathogenesis of IBD and provides a theoretical basis for the personalized approaches that modulate the gut microbiota to treat IBD.
Keywords: IBD; gut microbiota; inflammatory bowel disease; metabolite; treatment.
Copyright © 2022 Qiu, Ishimoto, Fu, Zhang, Zhang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Aden K., Rehman A., Waschina S., Pan W. H., Walker A., Lucio M., et al. . (2019). Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology 157 (5), 1279–1292.e1211. doi: 10.1053/j.gastro.2019.07.025 - DOI - PubMed
-
- Alexeev E. E., Lanis J. M., Kao D. J., Campbell E. L., Kelly C. J., Battista K. D., et al. . (2018). Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis Through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 188 (5), 1183–1194. doi: 10.1016/j.ajpath.2018.01.011 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

