Comprehensive Analyses of the Immunological and Prognostic Roles of an IQGAP3AR/let-7c-5p/IQGAP3 Axis in Different Types of Human Cancer
- PMID: 35274003
- PMCID: PMC8902246
- DOI: 10.3389/fmolb.2022.763248
Comprehensive Analyses of the Immunological and Prognostic Roles of an IQGAP3AR/let-7c-5p/IQGAP3 Axis in Different Types of Human Cancer
Abstract
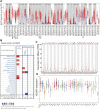
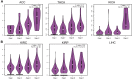
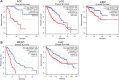
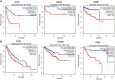
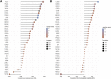
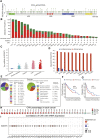
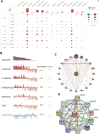
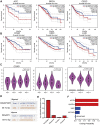
IQ motif containing GTPase-activating protein 3 (IQGAP3) is a member of the Rho family of guanosine-5'-triphosphatases (GTPases). IQGAP3 plays a crucial part in the development and progression of several types of cancer. However, the prognostic, upstream-regulatory, and immunological roles of IQGAP3 in human cancer types are not known. We found that IQGAP3 expression was increased in different types of human cancer. The high expression of IQGAP3 was correlated with tumor stage, lymph node metastasis, and a poor prognosis in diverse types of human cancer. The DNA methylation of IQGAP3 was highly and negatively correlated with IQGAP3 expression in diverse cancer types. High DNA methylation in IQGAP3 was correlated with better overall survival in human cancer types. High mRNA expression of IQGAP3 was associated with tumor mutational burden, microsatellite instability, immune cell infiltration, and immune modulators. Analyses of signaling pathway enrichment showed that IQGAP3 was involved in the cell cycle. IQGAP3 expression was associated with sensitivity to a wide array of drugs in cancer cells lines. We revealed that polypyrimidine tract-binding protein 1 (PTBP1) and an IQGAP3-associated lncRNA (IQGAP3AR)/let-7c-5p axis were potential regulations for IQGAP3 expression. We provided the first evidence to show that an IQGAP3AR/let-7c-5p/IQGAP3 axis has indispensable roles in the progression and immune response in different types of human cancer.
Keywords: IQGAP3; human cancer; immunotherapy; let-7c-5p; prognosis.
Copyright © 2022 Yuan, Jiang, Tang, Yang, Wang, Zhang and Duan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials

