Statelets: Capturing recurrent transient variations in dynamic functional network connectivity
- PMID: 35274791
- PMCID: PMC9057100
- DOI: 10.1002/hbm.25799
Statelets: Capturing recurrent transient variations in dynamic functional network connectivity
Abstract
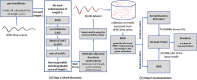
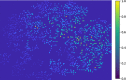
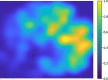
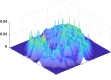
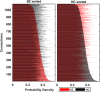
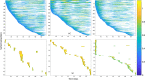
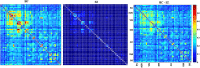
Dynamic functional network connectivity (dFNC) analysis is a widely used approach for capturing brain activation patterns, connectivity states, and network organization. However, a typical sliding window plus clustering (SWC) approach for analyzing dFNC models the system through a fixed sequence of connectivity states. SWC assumes connectivity patterns span throughout the brain, but they are relatively spatially constrained and temporally short-lived in practice. Thus, SWC is neither designed to capture transient dynamic changes nor heterogeneity across subjects/time. We propose a state-space time series summarization framework called "statelets" to address these shortcomings. It models functional connectivity dynamics at fine-grained timescales, adapting time series motifs to changes in connectivity strength, and constructs a concise yet informative representation of the original data that conveys easily comprehensible information about the phenotypes. We leverage the earth mover distance in a nonstandard way to handle scale differences and utilize kernel density estimation to build a probability density profile for local motifs. We apply the framework to study dFNC of patients with schizophrenia (SZ) and healthy control (HC). Results demonstrate SZ subjects exhibit reduced modularity in their brain network organization relative to HC. Statelets in the HC group show an increased recurrence across the dFNC time-course compared to the SZ. Analyzing the consistency of the connections across time reveals significant differences within visual, sensorimotor, and default mode regions where HC subjects show higher consistency than SZ. The introduced approach also enables handling dynamic information in cross-modal and multimodal applications to study healthy and disordered brains.
Keywords: dynamic functional network connectivity; earthmover distance; kernel density estimator; resting-state MRI; schizophrenia; time series motifs summarization.
© 2022 The Authors. Human Brain Mapping published by Wiley Periodicals LLC.
Conflict of interest statement
There is no conflict of interest.
Figures







References
-
- Ahmad, I. A. , & Amezziane, M. (2007). A general and fast convergent bandwidth selection method of kernel estimator. Journal of Nonparametric Statistics, 19(4–5), 165–187.
-
- Ahmad, S. , Taskaya‐Temizel, T. , & Ahmad, K. (2004). Summarizing time series: Learning patterns in ‘volatile’ series. In International conference on intelligent data engineering and automated learning. Exeter, England: Springer.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

