DLC-1 facilitates germ granule assembly in Caenorhabditis elegans embryo
- PMID: 35274966
- PMCID: PMC9282019
- DOI: 10.1091/mbc.E21-05-0275
DLC-1 facilitates germ granule assembly in Caenorhabditis elegans embryo
Abstract
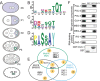
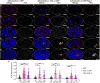
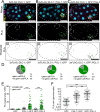
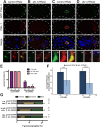
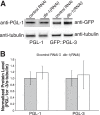
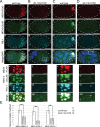
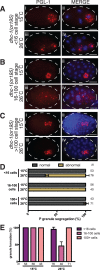
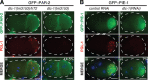
Germ granules are cytoplasmic assemblies of RNA-binding proteins (RBPs) required for germ cell development and fertility. During the first four cell divisions of the Caenorhabditis elegans zygote, regulated assembly of germ (P) granules leads to their selective segregation to the future germ cell. Here we investigate the role of DLC-1, a hub protein implicated in stabilization and function of diverse protein complexes, in maintaining P granule integrity. We find that DLC-1 directly interacts with several core P granule proteins, predominantly during embryogenesis. The loss of dlc-1 disrupts assembly of P granule components into phase-separated organelles in the embryos, regardless of whether or not DLC-1 directly interacts with these proteins. Finally, we infer that P granule dispersal in the absence of dlc-1 is likely independent of DLC-1's function as a subunit of the dynein motor and does not result from a loss of cell polarity.
Figures

Similar articles
-
Dynein Light Chain DLC-1 Facilitates the Function of the Germline Cell Fate Regulator GLD-1 in Caenorhabditis elegans.Genetics. 2019 Feb;211(2):665-681. doi: 10.1534/genetics.118.301617. Epub 2018 Dec 3. Genetics. 2019. PMID: 30509955 Free PMC article.
-
The Stress Granule RNA-Binding Protein TIAR-1 Protects Female Germ Cells from Heat Shock in Caenorhabditis elegans.G3 (Bethesda). 2016 Apr 7;6(4):1031-47. doi: 10.1534/g3.115.026815. G3 (Bethesda). 2016. PMID: 26865701 Free PMC article.
-
PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans.J Cell Biol. 2011 Mar 21;192(6):929-37. doi: 10.1083/jcb.201010106. Epub 2011 Mar 14. J Cell Biol. 2011. PMID: 21402787 Free PMC article.
-
Germ granules and gene regulation in the Caenorhabditis elegans germline.Genetics. 2022 Mar 3;220(3):iyab195. doi: 10.1093/genetics/iyab195. Genetics. 2022. PMID: 35239965 Free PMC article. Review.
-
Connecting the Dots: Linking Caenorhabditis elegans Small RNA Pathways and Germ Granules.Trends Cell Biol. 2021 May;31(5):387-401. doi: 10.1016/j.tcb.2020.12.012. Epub 2021 Jan 29. Trends Cell Biol. 2021. PMID: 33526340 Review.
Cited by
-
Disruption of C. elegans embryonic P granules upon dlc-1(RNAi) is not associated with P granule component loss.MicroPubl Biol. 2022 Dec 6;2022:10.17912/micropub.biology.000700. doi: 10.17912/micropub.biology.000700. eCollection 2022. MicroPubl Biol. 2022. PMID: 36568482 Free PMC article.
-
CED-6/GULP and components of the clathrin-mediated endocytosis machinery act redundantly to correctly display CED-1 on the cell membrane in Caenorhabditis elegans.G3 (Bethesda). 2024 Jul 8;14(7):jkae088. doi: 10.1093/g3journal/jkae088. G3 (Bethesda). 2024. PMID: 38696649 Free PMC article.
-
COP9 signalosome component CSN-5 stabilizes PUF proteins FBF-1 and FBF-2 in Caenorhabditis elegans germline stem and progenitor cells.Genetics. 2024 May 7;227(1):iyae033. doi: 10.1093/genetics/iyae033. Genetics. 2024. PMID: 38427913 Free PMC article.
References
-
- Bagchi S, Fredriksson R, Wallén-Mackenzie Å (2015). In situ proximity ligation assay (PLA). Methods Mol Biol 1318, 149–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

