Septins guide noncentrosomal microtubules to promote focal adhesion disassembly in migrating cells
- PMID: 35274967
- PMCID: PMC9282018
- DOI: 10.1091/mbc.E21-06-0334
Septins guide noncentrosomal microtubules to promote focal adhesion disassembly in migrating cells
Abstract
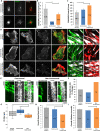
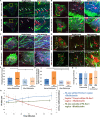
Endothelial cell migration is critical for vascular angiogenesis and is compromised to facilitate tumor metastasis. The migratory process requires the coordinated assembly and disassembly of focal adhesions (FA), actin, and microtubules (MT). MT dynamics at FAs deliver vesicular cargoes and enhance actomyosin contractility to promote FA turnover and facilitate cell advance. Noncentrosomal (NC) MTs regulate FA dynamics and are sufficient to drive cell polarity, but how NC MTs target FAs to control FA turnover is not understood. Here, we show that Rac1 induces the assembly of FA-proximal septin filaments that promote NC MT growth into FAs and inhibit mitotic centromere-associated kinesin (MCAK)-associated MT disassembly, thereby maintaining intact MT plus ends proximal to FAs. Septin-associated MT rescue is coupled with accumulation of Aurora-A kinase and cytoplasmic linker-associated protein (CLASP) localization to the MT between septin and FAs. In this way, NC MTs are strategically positioned to undergo MCAK- and CLASP-regulated bouts of assembly and disassembly into FAs, thereby regulating FA turnover and cell migration.
Figures






Similar articles
-
Adenomatous polyposis coli nucleates actin assembly to drive cell migration and microtubule-induced focal adhesion turnover.J Cell Biol. 2017 Sep 4;216(9):2859-2875. doi: 10.1083/jcb.201702007. Epub 2017 Jun 29. J Cell Biol. 2017. PMID: 28663347 Free PMC article.
-
Rac1 and Aurora A regulate MCAK to polarize microtubule growth in migrating endothelial cells.J Cell Biol. 2014 Jul 7;206(1):97-112. doi: 10.1083/jcb.201401063. J Cell Biol. 2014. PMID: 25002679 Free PMC article.
-
The compass to follow: Focal adhesion turnover.Curr Opin Cell Biol. 2023 Feb;80:102152. doi: 10.1016/j.ceb.2023.102152. Epub 2023 Feb 14. Curr Opin Cell Biol. 2023. PMID: 36796142 Review.
-
The role of APC-mediated actin assembly in microtubule capture and focal adhesion turnover.J Cell Biol. 2019 Oct 7;218(10):3415-3435. doi: 10.1083/jcb.201904165. Epub 2019 Aug 30. J Cell Biol. 2019. PMID: 31471457 Free PMC article.
-
Microtubules in cell migration.Essays Biochem. 2019 Oct 31;63(5):509-520. doi: 10.1042/EBC20190016. Essays Biochem. 2019. PMID: 31358621 Free PMC article. Review.
Cited by
-
Pharmacological inhibition of Septins with Forchlorfenuron attenuates thrombus formation in experimental thrombotic mice models with modulating multiple signaling pathways in platelets.J Adv Res. 2025 Jul;73:517-533. doi: 10.1016/j.jare.2024.08.006. Epub 2024 Aug 5. J Adv Res. 2025. PMID: 39111626 Free PMC article.
-
Septin and actin contributions to endothelial cell-cell junctions and monolayer integrity.Cytoskeleton (Hoboken). 2023 Jul-Aug;80(7-8):228-241. doi: 10.1002/cm.21732. Epub 2022 Oct 17. Cytoskeleton (Hoboken). 2023. PMID: 36205643 Free PMC article. Review.
-
ECM-dependent regulation of septin 7 in focal adhesions promotes mechanosensing and functional response in fibroblasts.iScience. 2024 Nov 9;27(12):111355. doi: 10.1016/j.isci.2024.111355. eCollection 2024 Dec 20. iScience. 2024. PMID: 39650732 Free PMC article.
-
The role of activated androgen receptor in cofilin phospho-regulation depends on the molecular subtype of TNBC cell line and actin assembly dynamics.PLoS One. 2022 Dec 30;17(12):e0279746. doi: 10.1371/journal.pone.0279746. eCollection 2022. PLoS One. 2022. PMID: 36584207 Free PMC article.
-
Multimodal illumination platform for 3D single-molecule super-resolution imaging throughout mammalian cells.Biomed Opt Express. 2024 Apr 16;15(5):3050-3063. doi: 10.1364/BOE.521362. eCollection 2024 May 1. Biomed Opt Express. 2024. PMID: 38855669 Free PMC article.
References
-
- Akhmanova A, Hoogengraad CC (2015). Microtubule minus-end-targeting proteins. Curr Biol 25, R162–R171. - PubMed
-
- Akhmanova A, Steinmetz MO (2010). Microtubule +TIPs at a glance. J Cell Sci 123, 3415–3419. - PubMed
-
- Aoki K, Nakamura T, Matsuda M (2004). Spatio-temporal regulation of Rac1 and Cdc42 activity during nerve growth factor-induced neurite outgrowth in PC12 cells. J Biol Chem 279, 713–719. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

