Single-molecule Taq DNA polymerase dynamics
- PMID: 35275726
- PMCID: PMC8916733
- DOI: 10.1126/sciadv.abl3522
Single-molecule Taq DNA polymerase dynamics
Abstract
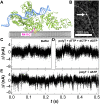
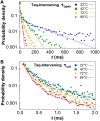
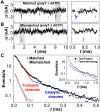
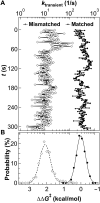
Taq DNA polymerase functions at elevated temperatures with fast conformational dynamics-regimes previously inaccessible to mechanistic, single-molecule studies. Here, single-walled carbon nanotube transistors recorded the motions of Taq molecules processing matched or mismatched template-deoxynucleotide triphosphate pairs from 22° to 85°C. By using four enzyme orientations, the whole-enzyme closures of nucleotide incorporations were distinguished from more rapid, 20-μs closures of Taq's fingers domain testing complementarity and orientation. On average, one transient closure was observed for every nucleotide binding event; even complementary substrate pairs averaged five transient closures between each catalytic incorporation at 72°C. The rate and duration of the transient closures and the catalytic events had almost no temperature dependence, leaving all of Taq's temperature sensitivity to its rate-determining open state.
Figures

References
-
- G. Maga, in Reference Module in Biomedical Sciences (Elsevier Inc., 2019), pp. 376–378.
-
- Aschenbrenner J., Marx A., DNA polymerases and biotechnological applications. Curr. Opin. Biotechnol. 48, 187–195 (2017). - PubMed
-
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A., Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491 (1988). - PubMed
-
- Eom S. H., Wang J., Steitz T. A., Structure of Taq polymerase with DNA at the polymerase active site. Nature 382, 278–281 (1996). - PubMed

