H-ABC- and dystonia-causing TUBB4A mutations show distinct pathogenic effects
- PMID: 35275727
- PMCID: PMC8916731
- DOI: 10.1126/sciadv.abj9229
H-ABC- and dystonia-causing TUBB4A mutations show distinct pathogenic effects
Abstract
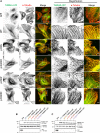
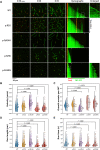
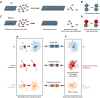
Mutations in the brain-specific β-tubulin 4A (TUBB4A) gene cause a broad spectrum of diseases, ranging from dystonia (DYT-TUBB4A) to hypomyelination with atrophy of the basal ganglia and cerebellum (H-ABC). Currently, the mechanisms of how TUBB4A variants lead to this pleiotropic manifestation remain elusive. Here, we investigated whether TUBB4A mutations causing either DYT-TUBB4A (p.R2G and p.Q424H) or H-ABC (p.R2W and p.D249N) exhibit differential effects at the molecular and cellular levels. Using live-cell imaging of disease-relevant oligodendrocytes and total internal reflection fluorescence microscopy of whole-cell lysates, we observed divergent impact on microtubule polymerization and microtubule integration, partially reflecting the observed pleiotropy. Moreover, in silico simulations demonstrated that the mutants rarely adopted a straight heterodimer conformation in contrast to wild type. In conclusion, for most of the examined variants, we deciphered potential molecular disease mechanisms that may lead to the diverse clinical manifestations and phenotype severity across and within each TUBB4A-related disease.
Figures



References
-
- Heinzen E. L., Swoboda K. J., Hitomi Y., Gurrieri F., Nicole S., de Vries B., Tiziano F. D., Fontaine B., Walley N. M., Heavin S., Panagiotakaki E.; European Alternating Hemiplegia of Childhood (AHC) Genetics Consortium; Biobanca e Registro Clinico per l’Emiplegia Alternante (I.B.AHC) Consortium; European Network for Research on Alternating Hemiplegia (ENRAH) for Small and Medium-sized Enterpriese (SMEs) Consortium, Fiori S., Abiusi E., Pietro L. D., Sweney M. T., Newcomb T. M., Viollet L., Huff C., Jorde L. B., Reyna S. P., Murphy K. J., Shianna K. V., Gumbs C. E., Little L., Silver K., Ptáček L. J., Haan J., Ferrari M. D., Bye A. M., Herkes G. K., Whitelaw C. M., Webb D., Lynch B. J., Uldall P., King M. D., Scheffer I. E., Neri G., Arzimanoglou A., van den Maagdenberg A. M. J. M., Sisodiya S. M., Mikati M. A., Goldstein D. B., De novo mutations in ATP1A3 cause alternating hemiplegia of childhood. Nat Genet. 44, 1030–1034 (2012). - PMC - PubMed
-
- Rosewich H., Thiele H., Ohlenbusch A., Maschke U., Altmüller J., Frommolt P., Zirn B., Ebinger F., Siemes H., Nürnberg P., Brockmann K., Gärtner J., Heterozygous de-novo mutations in ATP1A3 in patients with alternating hemiplegia of childhood: A whole-exome sequencing gene-identification study. Lancet Neurol. 11, 764–773 (2012). - PubMed
-
- de Carvalho Aguiar P., Sweadner K. J., Penniston J. T., Zaremba J., Liu L., Caton M., Linazasoro G., Borg M., Tijssen M. A. J., Bressman S. B., Dobyns W. B., Brashear A., Ozelius L. J., Mutations in the Na+/K+ -ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron 43, 169–175 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

