Diversity and Evolution of Pigment Types in Marine Synechococcus Cyanobacteria
- PMID: 35276007
- PMCID: PMC8995045
- DOI: 10.1093/gbe/evac035
Diversity and Evolution of Pigment Types in Marine Synechococcus Cyanobacteria
Abstract
Synechococcus cyanobacteria are ubiquitous and abundant in the marine environment and contribute to an estimated 16% of the ocean net primary productivity. Their light-harvesting complexes, called phycobilisomes (PBS), are composed of a conserved allophycocyanin core, from which radiates six to eight rods with variable phycobiliprotein and chromophore content. This variability allows Synechococcus cells to optimally exploit the wide variety of spectral niches existing in marine ecosystems. Seven distinct pigment types or subtypes have been identified so far in this taxon based on the phycobiliprotein composition and/or the proportion of the different chromophores in PBS rods. Most genes involved in their biosynthesis and regulation are located in a dedicated genomic region called the PBS rod region. Here, we examine the variability of gene content and organization of this genomic region in a large set of sequenced isolates and natural populations of Synechococcus representative of all known pigment types. All regions start with a tRNA-PheGAA and some possess mobile elements for DNA integration and site-specific recombination, suggesting that their genomic variability relies in part on a "tycheposon"-like mechanism. Comparison of the phylogenies obtained for PBS and core genes revealed that the evolutionary history of PBS rod genes differs from the core genome and is characterized by the co-existence of different alleles and frequent allelic exchange. We propose a scenario for the evolution of the different pigment types and highlight the importance of incomplete lineage sorting in maintaining a wide diversity of pigment types in different Synechococcus lineages despite multiple speciation events.
Keywords: cyanobacteria; genomic island; lateral gene transfer; phycobiliprotein; phycobilisome; tycheposon.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures





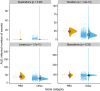
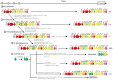
Similar articles
-
Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study.Genome Biol. 2007;8(12):R259. doi: 10.1186/gb-2007-8-12-r259. Genome Biol. 2007. PMID: 18062815 Free PMC article.
-
Characterization of Molecular Diversity and Organization of Phycobilisomes in Thermophilic Cyanobacteria.Int J Mol Sci. 2023 Mar 15;24(6):5632. doi: 10.3390/ijms24065632. Int J Mol Sci. 2023. PMID: 36982707 Free PMC article.
-
Development of a targeted metagenomic approach to study a genomic region involved in light harvesting in marine Synechococcus.FEMS Microbiol Ecol. 2014 May;88(2):231-49. doi: 10.1111/1574-6941.12285. Epub 2014 Feb 24. FEMS Microbiol Ecol. 2014. PMID: 24862161
-
How can Phycobilisome, the unique light harvesting system in certain algae working highly efficiently: The connection in between structures and functions.Prog Biophys Mol Biol. 2024 Jan;186:39-52. doi: 10.1016/j.pbiomolbio.2023.11.005. Epub 2023 Nov 28. Prog Biophys Mol Biol. 2024. PMID: 38030044 Review.
-
Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies.Trends Microbiol. 2002 Mar;10(3):134-42. doi: 10.1016/s0966-842x(02)02319-3. Trends Microbiol. 2002. PMID: 11864823 Review.
Cited by
-
Differential acclimation kinetics of the two forms of type IV chromatic acclimaters occurring in marine Synechococcus cyanobacteria.Front Microbiol. 2024 Feb 16;15:1349322. doi: 10.3389/fmicb.2024.1349322. eCollection 2024. Front Microbiol. 2024. PMID: 38435691 Free PMC article.
-
The phycoerythrobilin isomerization activity of MpeV in Synechococcus sp. WH8020 is prevented by the presence of a histidine at position 141 within its phycoerythrin-I β-subunit substrate.Front Microbiol. 2022 Nov 15;13:1011189. doi: 10.3389/fmicb.2022.1011189. eCollection 2022. Front Microbiol. 2022. PMID: 36458192 Free PMC article.
-
Genomic Insights on the Carbon-Negative Workhorse: Systematical Comparative Genomic Analysis on 56 Synechococcus Strains.Bioengineering (Basel). 2023 Nov 18;10(11):1329. doi: 10.3390/bioengineering10111329. Bioengineering (Basel). 2023. PMID: 38002453 Free PMC article.
-
Highly Diverse Synechococcus Pigment Types in the Eastern Indian Ocean.Front Microbiol. 2022 Feb 25;13:806390. doi: 10.3389/fmicb.2022.806390. eCollection 2022. Front Microbiol. 2022. PMID: 35283844 Free PMC article.
-
Phycobilisomes and Phycobiliproteins in the Pigment Apparatus of Oxygenic Photosynthetics: From Cyanobacteria to Tertiary Endosymbiosis.Int J Mol Sci. 2023 Jan 24;24(3):2290. doi: 10.3390/ijms24032290. Int J Mol Sci. 2023. PMID: 36768613 Free PMC article. Review.
References
-
- Abby S, Daubin V. 2007. Comparative genomics and the evolution of prokaryotes. Trends Microbiol. 15:135–141. - PubMed
-
- Adir N. 2005. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth Res. 85:15–32. - PubMed
-
- Ahlgren NA, Belisle BS, Lee MD. 2019. Genomic mosaicism underlies the adaptation of marine Synechococcus ecotypes to distinct oceanic iron niches. Environ Microbiol. 22:1801–1815. - PubMed
-
- Antoine D, et al. . 2008. Assessment of uncertainty in the ocean reflectance determined by three satellite ocean color sensors (MERIS, SeaWiFS and MODIS-A) at an offshore site in the Mediterranean Sea (BOUSSOLE project). J Geophys Res. 113:C07013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

