Molecular Formula Prediction for Chemical Filtering of 3D OrbiSIMS Datasets
- PMID: 35276049
- PMCID: PMC8943605
- DOI: 10.1021/acs.analchem.1c04898
Molecular Formula Prediction for Chemical Filtering of 3D OrbiSIMS Datasets
Abstract
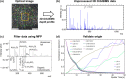
Modern mass spectrometry techniques produce a wealth of spectral data, and although this is an advantage in terms of the richness of the information available, the volume and complexity of data can prevent a thorough interpretation to reach useful conclusions. Application of molecular formula prediction (MFP) to produce annotated lists of ions that have been filtered by their elemental composition and considering structural double bond equivalence are widely used on high resolving power mass spectrometry datasets. However, this has not been applied to secondary ion mass spectrometry data. Here, we apply this data interpretation approach to 3D OrbiSIMS datasets, testing it for a series of increasingly complex samples. In an organic on inorganic sample, we successfully annotated the organic contaminant overlayer separately from the substrate. In a more challenging purely organic human serum sample we filtered out both proteins and lipids based on elemental compositions, 226 different lipids were identified and validated using existing databases, and we assigned amino acid sequences of abundant serum proteins including albumin, fibronectin, and transferrin. Finally, we tested the approach on depth profile data from layered carbonaceous engine deposits and annotated previously unidentified lubricating oil species. Application of an unsupervised machine learning method on filtered ions after performing MFP from this sample uniquely separated depth profiles of species, which were not observed when performing the method on the entire dataset. Overall, the chemical filtering approach using MFP has great potential in enabling full interpretation of complex 3D OrbiSIMS datasets from a plethora of material types.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

