Recent advances in the developmental origin of neuroblastoma: an overview
- PMID: 35277192
- PMCID: PMC8915499
- DOI: 10.1186/s13046-022-02281-w
Recent advances in the developmental origin of neuroblastoma: an overview
Abstract
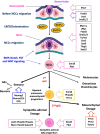
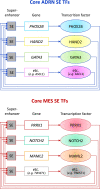
Neuroblastoma (NB) is a pediatric tumor that originates from neural crest-derived cells undergoing a defective differentiation due to genomic and epigenetic impairments. Therefore, NB may arise at any final site reached by migrating neural crest cells (NCCs) and their progeny, preferentially in the adrenal medulla or in the para-spinal ganglia.NB shows a remarkable genetic heterogeneity including several chromosome/gene alterations and deregulated expression of key oncogenes that drive tumor initiation and promote disease progression.NB substantially contributes to childhood cancer mortality, with a survival rate of only 40% for high-risk patients suffering chemo-resistant relapse. Hence, NB remains a challenge in pediatric oncology and the need of designing new therapies targeted to specific genetic/epigenetic alterations become imperative to improve the outcome of high-risk NB patients with refractory disease or chemo-resistant relapse.In this review, we give a broad overview of the latest advances that have unraveled the developmental origin of NB and its complex epigenetic landscape.Single-cell RNA sequencing with spatial transcriptomics and lineage tracing have identified the NCC progeny involved in normal development and in NB oncogenesis, revealing that adrenal NB cells transcriptionally resemble immature neuroblasts or their closest progenitors. The comparison of adrenal NB cells from patients classified into risk subgroups with normal sympatho-adrenal cells has highlighted that tumor phenotype severity correlates with neuroblast differentiation grade.Transcriptional profiling of NB tumors has identified two cell identities that represent divergent differentiation states, i.e. undifferentiated mesenchymal (MES) and committed adrenergic (ADRN), able to interconvert by epigenetic reprogramming and to confer intra-tumoral heterogeneity and high plasticity to NB.Chromatin immunoprecipitation sequencing has disclosed the existence of two super-enhancers and their associated transcription factor networks underlying MES and ADRN identities and controlling NB gene expression programs.The discovery of NB-specific regulatory circuitries driving oncogenic transformation and maintaining the malignant state opens new perspectives on the design of innovative therapies targeted to the genetic and epigenetic determinants of NB. Remodeling the disrupted regulatory networks from a dysregulated expression, which blocks differentiation and enhances proliferation, toward a controlled expression that prompts the most differentiated state may represent a promising therapeutic strategy for NB.
Keywords: Adrenergic; Core regulatory circuitries; Mesenchymal; Neural crest; Neuroblastoma; Transcription factors.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

