CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype
- PMID: 35277458
- PMCID: PMC8919471
- DOI: 10.1136/jitc-2021-004029
CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype
Abstract
Background: Platinum resistance is a major challenge in the clinical treatment of advanced ovarian cancer (OC). Accumulating evidence shows that the tumor-promotive M2 macrophage is linked to the limiting chemotherapy efficacy of multiple malignancies including OC. Circular RNAs (circRNAs) are a novel class of non-coding RNAs which function as the critical regulator in biological process of cancer. However, their impact on macrophage polarization and chemoresistance of OC remain unclear.
Methods: Platinum-resistant circRNAs were screened using circRNA deep sequencing and validated using in situ hybridization in OC tissues with or without platinum resistance. The role of circITGB6 in inducing cisplatin (CDDP) resistance was evaluated by clone formation, immunofluorescence and annexin V assays in vitro, and by intraperitoneal tumor model in vivo. The mechanism underlying circITGB6-mediated tumor-associated macrophage (TAM) polarization into M2 phenotype was investigated using RNA pull-down, luciferase reporter, electrophoretic mobility shift, RNA binding protein immunoprecipitation (RIP), ELISA and immunofluorescence assays.
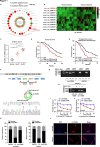
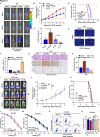
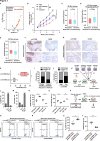
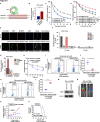
Results: We identified that a novel circRNA, circITGB6, robustly elevated in tumor tissues and serums from patients with OC with platinum resistance, was correlated with poor prognosis. circITGB6 overexpression promoted an M2 macrophage-dependent CDDP resistance in both vivo and vitro. Mechanistic research determined that circITGB6 directly interacted with IGF2BP2 and FGF9 mRNA to form a circITGB6/IGF2BP2/FGF9 RNA-protein ternary complex in the cytoplasm, thereby stabilizing FGF9 mRNA and inducing polarization of TAMs toward M2 phenotype. Importantly, blocking M2 macrophage polarization with an antisense oligonucleotide targeting circITGB6 markedly reversed the circITGB6-induced CDDP resistance of OC in vivo.
Conclusions: This study reveals a novel mechanism for platinum resistance in OC and demonstrates that circITGB6 may serve as a potential prognostic marker and a therapeutic target for patients with OC.
Keywords: macrophages; translational medical research; tumor microenvironment.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: There are no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
