The structural basis for regulation of the glutathione transporter Ycf1 by regulatory domain phosphorylation
- PMID: 35277487
- PMCID: PMC8917219
- DOI: 10.1038/s41467-022-28811-w
The structural basis for regulation of the glutathione transporter Ycf1 by regulatory domain phosphorylation
Abstract
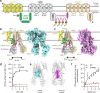
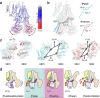
Yeast Cadmium Factor 1 (Ycf1) sequesters heavy metals and glutathione into the vacuole to counter cell stress. Ycf1 belongs to the ATP binding cassette C-subfamily (ABCC) of transporters, many of which are regulated by phosphorylation on intrinsically-disordered domains. The regulatory mechanism of phosphorylation is still poorly understood. Here, we report two cryo-EM structures of Ycf1 at 3.4 Å and 4.0 Å resolution in inward-facing open conformations that capture previously unobserved ordered states of the intrinsically disordered regulatory domain (R-domain). R-domain phosphorylation is clearly evident and induces a topology promoting electrostatic and hydrophobic interactions with Nucleotide Binding Domain 1 (NBD1) and the Lasso motif. These interactions stay constant between the structures and are related by rigid body movements of the NBD1/R-domain complex. Biochemical data further show R-domain phosphorylation reorganizes the Ycf1 architecture and is required for maximal ATPase activity. Together, we provide insights into how R-domains control ABCC transporter activity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Gueldry O, et al. Ycf1p-dependent Hg(II) detoxification in Saccharomyces cerevisiae. Eur. J. Biochem. 2003;270:2486–2496. - PubMed
-
- Szczypka MS, Wemmie JA, Moye-Rowley WS, Thiele DJ. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 1994;269:22853–22857. - PubMed
-
- Morgan B, et al. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 2013;9:119–125. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

