AC010973.2 promotes cell proliferation and is one of six stemness-related genes that predict overall survival of renal clear cell carcinoma
- PMID: 35277527
- PMCID: PMC8917182
- DOI: 10.1038/s41598-022-07070-1
AC010973.2 promotes cell proliferation and is one of six stemness-related genes that predict overall survival of renal clear cell carcinoma
Abstract
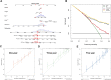
Extensive research indicates that tumor stemness promotes tumor progression. Nonetheless, the underlying roles of stemness-related genes in renal clear cell carcinoma (ccRCC) are unclear. Data used in bioinformatics analysis were downloaded from The Cancer Genome Atlas (TCGA) database. Moreover, the R software, SPSS, and GraphPad Prism 8 were used for mapping and statistical analysis. First, the stemness index of each patient was quantified using a machine learning algorithm. Subsequently, the differentially expressed genes between high and low stemness index were identified as stemness-related genes. Based on these genes, a stable and effective prognostic model was identified to predict the overall survival of patients using a random forest algorithm (Training cohort; 1-year AUC: 0.67; 3-year AUC: 0.79; 5-year AUC: 0.73; Validation cohort; 1-year AUC: 0.66; 3-year AUC: 0.71; 5-year AUC: 0.7). The model genes comprised AC010973.2, RNU6-125P, AP001209.2, Z98885.1, KDM5C-IT1, and AL021368.3. Due to its highest importance evaluated by randomforst analysis, the AC010973.2 gene was selected for further research. In vitro experiments demonstrated that AC010973.2 is highly expressed in ccRCC tissue and cell lines. Meanwhile, its knockdown could significantly inhibit the proliferation of ccRCC cells based on colony formation and CCK8 assays. In summary, our findings reveal that the stemness-related gene AC01097.3 is closely associated with the survival of patients. Besides, it remarkably promotes cell proliferation in ccRCC, hence a novel potential therapeutic target.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Alternative splicing associated with cancer stemness in kidney renal clear cell carcinoma.BMC Cancer. 2021 Jun 15;21(1):703. doi: 10.1186/s12885-021-08470-8. BMC Cancer. 2021. PMID: 34130646 Free PMC article.
-
ENAM gene associated with T classification and inhibits proliferation in renal clear cell carcinoma.Aging (Albany NY). 2021 Feb 3;13(5):7035-7051. doi: 10.18632/aging.202558. Epub 2021 Feb 3. Aging (Albany NY). 2021. PMID: 33539322 Free PMC article.
-
Identification of an independent immune-genes prognostic index for renal cell carcinoma.BMC Cancer. 2021 Jun 29;21(1):746. doi: 10.1186/s12885-021-08367-6. BMC Cancer. 2021. PMID: 34187413 Free PMC article.
-
Genome-wide screening and cohorts validation identifying novel lncRNAs as prognostic biomarkers for clear cell renal cell carcinoma.J Cell Biochem. 2020 Mar;121(3):2559-2570. doi: 10.1002/jcb.29478. Epub 2019 Oct 23. J Cell Biochem. 2020. PMID: 31646670
-
Prognosis of clear cell renal cell carcinoma (ccRCC) based on a six-lncRNA-based risk score: an investigation based on RNA-sequencing data.J Transl Med. 2019 Aug 23;17(1):281. doi: 10.1186/s12967-019-2032-y. J Transl Med. 2019. PMID: 31443717 Free PMC article.
Cited by
-
MEX3C as a potential target for hepatocellular carcinoma drug and immunity: combined therapy with Lenvatinib.BMC Cancer. 2023 Oct 12;23(1):967. doi: 10.1186/s12885-023-11320-4. BMC Cancer. 2023. PMID: 37828435 Free PMC article.
-
A novel PD-1/PD-L1 pathway molecular typing-related signature for predicting prognosis and the tumor microenvironment in breast cancer.Discov Oncol. 2023 May 8;14(1):59. doi: 10.1007/s12672-023-00669-4. Discov Oncol. 2023. PMID: 37154982 Free PMC article.
-
High expression of CCDC69 is correlated with immunotherapy response and protective effects on breast cancer.BMC Cancer. 2023 Oct 12;23(1):974. doi: 10.1186/s12885-023-11411-2. BMC Cancer. 2023. PMID: 37828454 Free PMC article.
-
TUBA1C is a potential new prognostic biomarker and promotes bladder urothelial carcinoma progression by regulating the cell cycle.BMC Cancer. 2023 Aug 1;23(1):716. doi: 10.1186/s12885-023-11209-2. BMC Cancer. 2023. PMID: 37528357 Free PMC article.
-
Second primary colorectal cancer in adults: a SEER analysis of incidence and outcomes.BMC Gastroenterol. 2023 Jul 26;23(1):253. doi: 10.1186/s12876-023-02893-2. BMC Gastroenterol. 2023. PMID: 37495987 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

