Computational identification of host genomic biomarkers highlighting their functions, pathways and regulators that influence SARS-CoV-2 infections and drug repurposing
- PMID: 35277538
- PMCID: PMC8915158
- DOI: 10.1038/s41598-022-08073-8
Computational identification of host genomic biomarkers highlighting their functions, pathways and regulators that influence SARS-CoV-2 infections and drug repurposing
Abstract
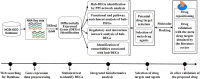
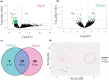
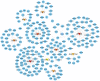
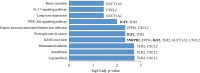
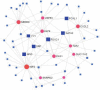
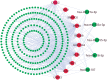
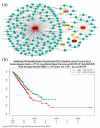
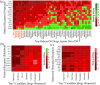
The pandemic threat of COVID-19 has severely destroyed human life as well as the economy around the world. Although, the vaccination has reduced the outspread, but people are still suffering due to the unstable RNA sequence patterns of SARS-CoV-2 which demands supplementary drugs. To explore novel drug target proteins, in this study, a transcriptomics RNA-Seq data generated from SARS-CoV-2 infection and control samples were analyzed. We identified 109 differentially expressed genes (DEGs) that were utilized to identify 10 hub-genes/proteins (TLR2, USP53, GUCY1A2, SNRPD2, NEDD9, IGF2, CXCL2, KLF6, PAG1 and ZFP36) by the protein-protein interaction (PPI) network analysis. The GO functional and KEGG pathway enrichment analyses of hub-DEGs revealed some important functions and signaling pathways that are significantly associated with SARS-CoV-2 infections. The interaction network analysis identified 5 TFs proteins and 6 miRNAs as the key regulators of hub-DEGs. Considering 10 hub-proteins and 5 key TFs-proteins as drug target receptors, we performed their docking analysis with the SARS-CoV-2 3CL protease-guided top listed 90 FDA approved drugs. We found Torin-2, Rapamycin, Radotinib, Ivermectin, Thiostrepton, Tacrolimus and Daclatasvir as the top ranked seven candidate drugs. We investigated their resistance performance against the already published COVID-19 causing top-ranked 11 independent and 8 protonated receptor proteins by molecular docking analysis and found their strong binding affinities, which indicates that the proposed drugs are effective against the state-of-the-arts alternatives independent receptor proteins also. Finally, we investigated the stability of top three drugs (Torin-2, Rapamycin and Radotinib) by using 100 ns MD-based MM-PBSA simulations with the two top-ranked proposed receptors (TLR2, USP53) and independent receptors (IRF7, STAT1), and observed their stable performance. Therefore, the proposed drugs might play a vital role for the treatment against different variants of SARS-CoV-2 infections.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Coronavirus disease 2019 (COVID-19) during pregnancy: prevalence of seroconversion, effect on maternal and perinatal outcomes and risk of vertical transmission.1 (2020).
-
- Fox S. SARS update. Infect. Med. 2003;20:269.
-
- WHO | Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. WHO (2015).
-
- Peiris JSM, Yuen KY, Osterhaus ADME, Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. - PubMed
-
- Leung GM, et al. The epidemiology of severe acute respiratory syndrome in the 2003 Hong Kong epidemic: An analysis of all 1755 patients. Ann. Intern. Med. 2004;141:662. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

