Understanding the functional role of membrane confinements in TNF-mediated signaling by multiscale simulations
- PMID: 35277586
- PMCID: PMC8917213
- DOI: 10.1038/s42003-022-03179-1
Understanding the functional role of membrane confinements in TNF-mediated signaling by multiscale simulations
Abstract
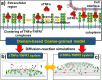
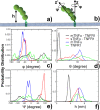
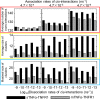
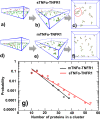
The interaction between TNFα and TNFR1 is essential in maintaining tissue development and immune responses. While TNFR1 is a cell surface receptor, TNFα exists in both soluble and membrane-bound forms. Interestingly, it was found that the activation of TNFR1-mediated signaling pathways is preferentially through the soluble form of TNFα, which can also induce the clustering of TNFR1 on plasma membrane of living cells. We developed a multiscale simulation framework to compare receptor clustering induced by soluble and membrane-bound ligands. Comparing with the freely diffusive soluble ligands, we hypothesize that the conformational dynamics of membrane-bound ligands are restricted, which affects the clustering of ligand-receptor complexes at cell-cell interfaces. Our simulation revealed that only small clusters can form if TNFα is bound on cell surface. In contrast, the clustering triggered by soluble TNFα is more dynamic, and the size of clusters is statistically larger. We therefore demonstrated the impact of membrane-bound ligand on dynamics of receptor clustering. Moreover, considering that larger TNFα-TNFR1 clusters is more likely to provide spatial platform for downstream signaling pathway, our studies offer new mechanistic insights about why the activation of TNFR1-mediated signaling pathways is not preferred by membrane-bound form of TNFα.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Dostert C, Grusdat M, Letellier E, Brenner D. The TNF family of ligands and receptors: communication modules in the immune system and beyond. Physiol. Rev. 2019;99:115–160. - PubMed
-
- Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

