Transposable element insertion: a hidden major source of domesticated phenotypic variation in Brassica rapa
- PMID: 35278263
- PMCID: PMC9241368
- DOI: 10.1111/pbi.13807
Transposable element insertion: a hidden major source of domesticated phenotypic variation in Brassica rapa
Abstract
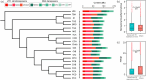
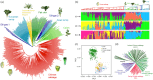
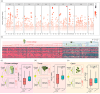
Transposable element (TE) is prevalent in plant genomes. However, studies on their impact on phenotypic evolution in crop plants are relatively rare, because systematically identifying TE insertions within a species has been a challenge. Here, we present a novel approach for uncovering TE insertion polymorphisms (TIPs) using pan-genome analysis combined with population-scale resequencing, and we adopt this pipeline to retrieve TIPs in a Brassica rapa germplasm collection. We found that 23% of genes within the reference Chiifu-401-42 genome harbored TIPs. TIPs tended to have large transcriptional effects, including modifying gene expression levels and altering gene structure by introducing new introns. Among 524 diverse accessions, TIPs broadly influenced genes related to traits and acted a crucial role in the domestication of B. rapa morphotypes. As examples, four specific TIP-containing genes were found to be candidates that potentially involved in various climatic conditions, promoting the formation of diverse vegetable crops in B. rapa. Our work reveals the hitherto hidden TIPs implicated in agronomic traits and highlights their widespread utility in studies of crop domestication.
Keywords: Brassica rapa; crop domestication; intraspecific diversification; pan-genome; transposable element insertion.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
Authors declare that they have no competing interests.
Figures




Similar articles
-
Genome resequencing and comparative variome analysis in a Brassica rapa and Brassica oleracea collection.Sci Data. 2016 Dec 20;3:160119. doi: 10.1038/sdata.2016.119. Sci Data. 2016. PMID: 27996963 Free PMC article.
-
Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa.Genome Biol. 2021 May 31;22(1):166. doi: 10.1186/s13059-021-02383-2. Genome Biol. 2021. PMID: 34059118 Free PMC article.
-
BrassicaTED - a public database for utilization of miniature transposable elements in Brassica species.BMC Res Notes. 2014 Jun 20;7:379. doi: 10.1186/1756-0500-7-379. BMC Res Notes. 2014. PMID: 24948109 Free PMC article.
-
The important contribution of transposable elements to phenotypic variation and evolution.Curr Opin Plant Biol. 2022 Feb;65:102140. doi: 10.1016/j.pbi.2021.102140. Epub 2021 Dec 6. Curr Opin Plant Biol. 2022. PMID: 34883307 Review.
-
Causes of Mutation Rate Variability in Plant Genomes.Annu Rev Plant Biol. 2023 May 22;74:751-775. doi: 10.1146/annurev-arplant-070522-054109. Epub 2023 Mar 8. Annu Rev Plant Biol. 2023. PMID: 36889008 Review.
Cited by
-
Genome-wide analysis of transposable elements and satellite DNA in Humulus scandens, a dioecious plant with XX/XY1Y2 chromosomes.Front Plant Sci. 2023 Oct 16;14:1230250. doi: 10.3389/fpls.2023.1230250. eCollection 2023. Front Plant Sci. 2023. PMID: 37908838 Free PMC article.
-
Look4LTRs: a Long terminal repeat retrotransposon detection tool capable of cross species studies and discovering recently nested repeats.Mob DNA. 2024 Apr 16;15(1):8. doi: 10.1186/s13100-024-00317-w. Mob DNA. 2024. PMID: 38627766 Free PMC article.
-
Multi-integrated genomic data for Passiflora foetida provides insights into genome size evolution and floral development in Passiflora.Mol Hortic. 2023 Dec 18;3(1):27. doi: 10.1186/s43897-023-00076-x. Mol Hortic. 2023. PMID: 38105261 Free PMC article.
-
Phased secondary small interfering RNAs in Camellia sinensis var. assamica.NAR Genom Bioinform. 2023 Nov 24;5(4):lqad103. doi: 10.1093/nargab/lqad103. eCollection 2023 Dec. NAR Genom Bioinform. 2023. PMID: 38025046 Free PMC article.
-
Graphical pangenomics-enabled characterization of structural variant impact on gene expression in Brassica napus.Theor Appl Genet. 2025 Apr 3;138(4):91. doi: 10.1007/s00122-025-04867-2. Theor Appl Genet. 2025. PMID: 40178590 Free PMC article.
References
-
- Baduel, P. , Quadrana, L. and Colot, V. (2021) Efficient detection of transposable element insertion polymorphisms between genomes using short‐read sequencing data. Methods Mol. Biol. 2250, 157–169. - PubMed
-
- Bayer, P.E. , Golicz, A.A. , Scheben, A. , Batley, J. and Edwards, D. (2020) Plant pan‐genomes are the new reference. Nat. Plants, 6, 914–920. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

