Potential inhibitor for blocking binding between ACE2 and SARS-CoV-2 spike protein with mutations
- PMID: 35279013
- PMCID: PMC8906167
- DOI: 10.1016/j.biopha.2022.112802
Potential inhibitor for blocking binding between ACE2 and SARS-CoV-2 spike protein with mutations
Abstract
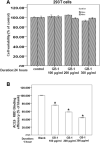
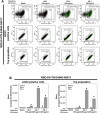
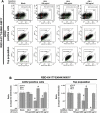
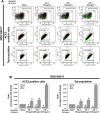
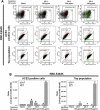
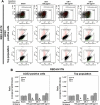
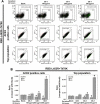
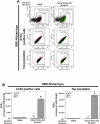
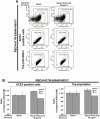
At the time of writing, more than 440 million confirmed coronavirus disease 2019 (COVID-19) cases and more than 5.97 million COVID-19 deaths worldwide have been reported by the World Health Organization since the start of the outbreak of the pandemic in Wuhan, China. During the COVID-19 pandemic, many variants of SARS-CoV-2 have arisen because of high mutation rates. N501Y, E484K, K417N, K417T, L452R and T478K in the receptor binding domain (RBD) region may increase the infectivity in several variants of SARS-CoV-2. In this study, we discovered that GB-1, developed from Chiehyuan herbal formula which obtained from Tian Shang Sheng Mu of Chiayi Puzi Peitian Temple, can inhibit the binding between ACE2 and RBD with Wuhan type, K417N-E484K-N501Y and L452R-T478K mutation. In addition, GB-1 inhibited the binding between ACE2 and RBD with a single mutation (E484K or N501Y), except the K417N mutation. In the compositions of GB-1, glycyrrhizic acid can inhibit the binding between ACE2 and RBD with Wuhan type, except K417N-E484K-N501Y mutation. Our results suggest that GB-1 could be a potential candidate for the prophylaxis of different variants of SARS-CoV-2 infection because of its inhibition of binding between ACE2 and RBD with different mutations (L452R-T478K, K417N-E484K-N501Y, N501Y or E484K).
Keywords: (+)-Catechin (Pubchem CID: 9064); COVID-19; GB-1; Glycyrrhizic acid; Glycyrrhizic acid (Pubchem CID: 14982); SARS-CoV-2; Spike protein.
Copyright © 2022 The Authors. Published by Elsevier Masson SAS.. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
GB-2 blocking the interaction between ACE2 and wild type and mutation of spike protein of SARS-CoV-2.Biomed Pharmacother. 2021 Oct;142:112011. doi: 10.1016/j.biopha.2021.112011. Epub 2021 Aug 5. Biomed Pharmacother. 2021. PMID: 34388530 Free PMC article.
-
Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19).2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 34033342 Free Books & Documents.
-
Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics.Elife. 2021 Aug 26;10:e70658. doi: 10.7554/eLife.70658. Elife. 2021. PMID: 34435953 Free PMC article.
-
Physicochemical effect of the N501Y, E484K/Q, K417N/T, L452R and T478K mutations on the SARS-CoV-2 spike protein RBD and its influence on agent fitness and on attributes developed by emerging variants of concern.Virology. 2022 Jul;572:44-54. doi: 10.1016/j.virol.2022.05.003. Epub 2022 May 12. Virology. 2022. PMID: 35580380 Free PMC article. Review.
-
Understanding Mutations in Human SARS-CoV-2 Spike Glycoprotein: A Systematic Review & Meta-Analysis.Viruses. 2023 Mar 27;15(4):856. doi: 10.3390/v15040856. Viruses. 2023. PMID: 37112836 Free PMC article.
Cited by
-
Research on the global trends of COVID-19 associated acute kidney injury: a bibliometric analysis.Ren Fail. 2024 Dec;46(1):2338484. doi: 10.1080/0886022X.2024.2338484. Epub 2024 Jun 4. Ren Fail. 2024. PMID: 38832469 Free PMC article. Review.
-
Glycyrrhizin and boswellic acids, the golden nutraceuticals: multitargeting for treatment of mild-moderate COVID-19 and prevention of post-COVID cognitive impairment.Inflammopharmacology. 2022 Dec;30(6):1977-1992. doi: 10.1007/s10787-022-01062-3. Epub 2022 Sep 22. Inflammopharmacology. 2022. PMID: 36136251 Free PMC article. Review.
-
Rational Design Problematics of Peptide Nucleic Acids as SARS-CoV-2 Inhibitors.ACS Omega. 2024 Jul 16;9(30):33000-33010. doi: 10.1021/acsomega.4c04023. eCollection 2024 Jul 30. ACS Omega. 2024. PMID: 39100288 Free PMC article.
-
The anti-SARS-CoV-2 effect and mechanism of Chiehyuan herbal oral protection solution.Heliyon. 2023 Jul 3;9(7):e17701. doi: 10.1016/j.heliyon.2023.e17701. eCollection 2023 Jul. Heliyon. 2023. PMID: 37483781 Free PMC article.
-
Nutrients, herbal bioactive derivatives and commensal microbiota as tools to lower the risk of SARS-CoV-2 infection.Front Nutr. 2023 Jun 1;10:1152254. doi: 10.3389/fnut.2023.1152254. eCollection 2023. Front Nutr. 2023. PMID: 37324739 Free PMC article. Review.
References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

