Vitamins D2 and D3 Have Overlapping But Different Effects on the Human Immune System Revealed Through Analysis of the Blood Transcriptome
- PMID: 35281034
- PMCID: PMC8908317
- DOI: 10.3389/fimmu.2022.790444
Vitamins D2 and D3 Have Overlapping But Different Effects on the Human Immune System Revealed Through Analysis of the Blood Transcriptome
Abstract
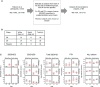
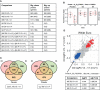
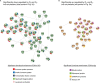
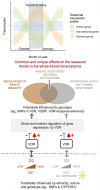
Vitamin D is best known for its role in maintaining bone health and calcium homeostasis. However, it also exerts a broad range of extra-skeletal effects on cellular physiology and on the immune system. Vitamins D2 and D3 share a high degree of structural similarity. Functional equivalence in their vitamin D-dependent effects on human physiology is usually assumed but has in fact not been well defined experimentally. In this study we seek to redress the gap in knowledge by undertaking an in-depth examination of changes in the human blood transcriptome following supplementation with physiological doses of vitamin D2 and D3. Our work extends a previously published randomized placebo-controlled trial that recruited healthy white European and South Asian women who were given 15 µg of vitamin D2 or D3 daily over 12 weeks in wintertime in the UK (Nov-Mar) by additionally determining changes in the blood transcriptome over the intervention period using microarrays. An integrated comparison of the results defines both the effect of vitamin D3 or D2 on gene expression, and any influence of ethnic background. An important aspect of this analysis was the focus on the changes in expression from baseline to the 12-week endpoint of treatment within each individual, harnessing the longitudinal design of the study. Whilst overlap in the repertoire of differentially expressed genes was present in the D2 or D3-dependent effects identified, most changes were specific to either one vitamin or the other. The data also pointed to the possibility of ethnic differences in the responses. Notably, following vitamin D3 supplementation, the majority of changes in gene expression reflected a down-regulation in the activity of genes, many encoding pathways of the innate and adaptive immune systems, potentially shifting the immune system to a more tolerogenic status. Surprisingly, gene expression associated with type I and type II interferon activity, critical to the innate response to bacterial and viral infections, differed following supplementation with either vitamin D2 or vitamin D3, with only vitamin D3 having a stimulatory effect. This study suggests that further investigation of the respective physiological roles of vitamin D2 and vitamin D3 is warranted.
Keywords: adaptive immunity; ethnicity; human transcriptome; immunomodulation; innate immunity; vitamin D supplementation; vitamin D2; vitamin D3.
Copyright © 2022 Durrant, Bucca, Hesketh, Möller-Levet, Tripkovic, Wu, Hart, Mathers, Elliott, Lanham-New and Smith.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

