Determining the Prognostic Value of Spliceosome-Related Genes in Hepatocellular Carcinoma Patients
- PMID: 35281269
- PMCID: PMC8907852
- DOI: 10.3389/fmolb.2022.759792
Determining the Prognostic Value of Spliceosome-Related Genes in Hepatocellular Carcinoma Patients
Abstract
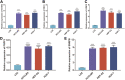
Background: The spliceosome plays an important role in mRNA alternative splicing and is aberrantly expressed in several tumors. However, the potential roles of spliceosome-related genes in the progression of hepatocellular carcinoma (HCC) remain poorly understood. Materials and Methods: Patient data were acquired from public databases. Expression differences and survival analyses were used to assess the importance of spliceosome-related genes in HCC prognosis. To explore the potential regulatory mechanisms of these genes, a protein-protein interaction network was constructed and screened using univariate and multivariate Cox regression and random forest analyses. This was used to create a five-gene prognostic model. The prognostic value and predictive power of the five-gene signature were assessed using the Kaplan-Meier and time-dependent receiver operating characteristic analyses in the training set. These results were further validated in an independent external set. To facilitate clinical application, a nomogram was prepared to predict the overall survival of HCC patients. The relative expression of five genes was detected using real-time quantitative polymerase chain reaction. Results: The analysis revealed that LSM1-7, SNRPB, SNRPD1-3, SNRPE, SNRPF, SNRPG, and SNRPN could be used as prognostic biomarkers in HCC patients. Moreover, the five-gene risk model could clearly distinguish between the high-and low-risk groups. Furthermore, the risk model was associated with the tumor mutation burden, immune cell infiltration of CD8+ T cells, natural killer T cells, M2 macrophages, and immune checkpoint inhibitors, which also demonstrated the predictive efficacy of this risk model in HCC immunotherapy. Conclusion: Spliceosome-related genes and the five-gene signature could serve as novel prognostic biomarkers for HCC patients, aiding clinical patient monitoring and follow-up.
Keywords: HCC; TMB; immune cell; prognostic genes; snRNP.
Copyright © 2022 Liu, Gu, Zhang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Berhane S., Toyoda H., Tada T., Kumada T., Kagebayashi C., Satomura S., et al. (2016). Role of the GALAD and BALAD-2 Serologic Models in Diagnosis of Hepatocellular Carcinoma and Prediction of Survival in Patients. Clin. Gastroenterol. Hepatol. 14 (6), 875–886. 10.1016/j.cgh.2015.12.042 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

